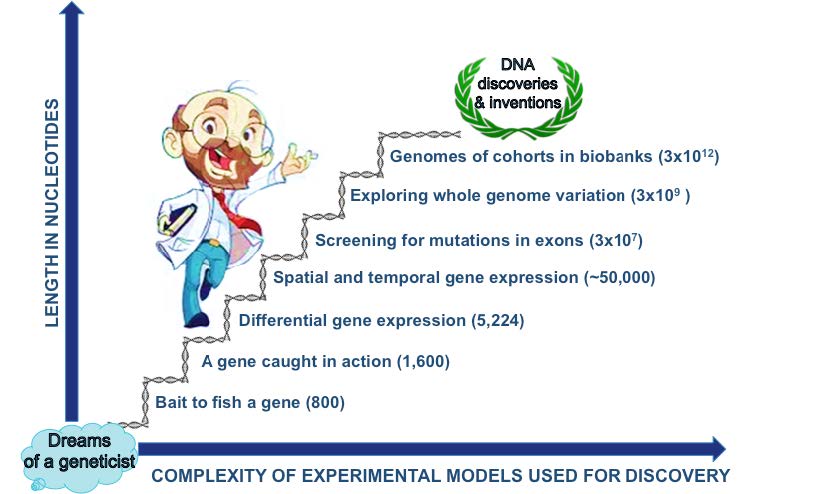
A Journey from Genes to Gene Families and on to the Whole Genome
Life sciences are coming back once again to dominate world wealth creation by harnessing the power of the ever-surprising scientific and technological breakthroughs. I have witnessed and participated in some of the most extraordinary ones: molecular cloning, polymerase chain reaction, the Human Genome Project, gene therapy, and personalized medicine, mainly. To ensure that this new era of accelerated discoveries continues, we need to engage the interest and curiosity of our youth and make them aware that a scientific career offers many opportunities to serve humanity while at the same time enjoying the wonders of an adventurous journey. This article tells the story of my personal voyage through the fascinating world of genetics, and of a series of discoveries and inventions that connect conventional concepts with more advanced aspects of genomics.
Keywords: cDNA; Gene Expression; Genome Mining; Cancer Genome; Biobanks
It was in my high school biology textbook in my hometown, a small Mexican village at the border with USA, that I first encountered genetics in a bewitching manner: “deoxyribonucleic acid holds the secret of life”, it read. Since biologist, James D. Watson, had identified its structure I entered the study of biology dreaming ‘there will come a day when I join those aspiring to reveal such secret’. Soon I realized that within biology it was in biochemistry where the nucleic acids were studied. Thus, I redirected my interest and efforts to become a biochemist, precisely owing to the nature of this mysterious molecule. To achieve my goal, I had to complement my biology curricula with extracurricular courses in chemistry, microbiology, and genetics. I had the wonderful opportunity to sit in the class of some amazing professors who told us the story of what was destined to be the discipline of molecular biology. I learned about the trail leading from Mendel to Watson and Crick, and was enchanted by the power of the biochemistry laboratory experimentation. The theoretical teaching was magnificently complemented by laboratory training, sitting as a volunteer in several non-curricular experimental advanced courses. These were usually offered during the summers and organized by an exceptional group of individuals including the most brilliant and academically ambitious students, led by also exceptional professors coming from prestigious universities in the country´s capital, who had agreed to nurture such ambition. In such courses, I was taught how to isolate and assay enzymes, to test for auxotrophies of different bacterial strains, and to chemically induce mutations in bacteria. But my encounter with DNA itself would have to wait until I entered the realm of the molecular biologists.
The teacher of the advanced biochemistry extracurricular course I attended had worked with bacterial RNA polymerase during his postdoctoral research at the University of California at Davis. A semester of continuous fights began because I questioned his frequent cancelation of classes. He retaliated by asking me tough questions about the reading material we were asked to prepare for each lesson. Since I slept with Lehninger´s textbook under my pillow, I always met his challenges. Eventually, by the end of the semester we agreed a cease-fire and subsequently a lasting friendship developed. At the end of the course he invited me to do my BSc thesis at his laboratory. While preparing the laboratory bench to grow bacteria to isolate RNA polymerase, the summer arrived, and I had decided to travel to the USA to attend an intensive English language course for international students held at the University of Houston. After a month I was desperate to return to biochemistry. The opportunity came as a volunteer in the Biochemistry Department of the MD Anderson Cancer Center, where for the rest of the afternoons of that memorable summer I learned how to isolate human placental nuclei and from them the long-sought DNA.
Adopting the placenta as my experimental model to enter the human molecular biology arena, turned out to be the most fortunate decision for a molecular biologist in a developing country. A good placenta weights about half a kilogram and is a generous source of human cells [1]. Besides DNA I also learned how to isolate total RNA and from it how to select by affinity chromatography on Oligo (dT)-cellulose the poly-A+ fraction which more or less equals to the messenger RNA. The need for more DNA or RNA was never a problem; I simply went to the delivery room at the Gynecology Department of a major local hospital, waited until the good news is given to the impatient father to be, and then went behind doors for a nurse to hand me the placenta that I immediately put on ice. Life in the laboratory was full of excitement with repeated cycles of isolating total human placental RNA, selecting the mRNA fraction, followed by the spectrophotometric quantification and electrophoretic characterization of both types of ribonucleic acids. Then using them in in vitro translation experiments that allowed us to identify the molecule responsible for the amazingly abundant placental mRNA and its derived protein, which turned out to be human placental lactogen (HPL, later renamed chorionic somatomammotropin or CSH) [2].
When I had thought that life in the laboratory could not be easier, I encountered a new and powerful technology that not only made life in the laboratory easier but also more productive: molecular cloning. I took advantage of it and isolated for the first time the full-length complementary DNA (cDNA) to the mRNA of HPL [3].
This hPL cDNA disclosed the molecular anatomy of one of the first human mRNAs discovered. Having known from the protein studies that the length of the HPL polypeptide was 22,000 daltons, we expected its mRNA to codify for a protein of about 190 amino acids. Indeed, we found an open reading frame of 651 codons counting from the initiation codon to the termination codon, which coded for a 190 amino-acid polypeptide having an amino-terminal hydrophobic extension of 26 amino acids typical of a secretion signal of proteins ending in the circulation. It also harbors some 30 extra nucleotides before the initiation codon (5´-end untranslated region or UTR) and some 100 extra in the other end after the termination codon (3´-end UTR), which are followed by an approximately 200-nucleotide poly-A tail. Therefore, a roughly 800 nucleotide-piece of DNA became my bait to catch a human gene.
And with the hPL cDNA clone came outstanding progress: the chromosomal localization of this gene locus as the first test for a newly invented technique developed in our laboratory (in situ hybridization); the discovery that the locus is comprised of five genes: three of hPL type and two of growth hormone (GH>) type; the confirmation that indeed the hPL-type genes in this locus are responsible in placenta for the amazing amounts of HPL; the surprising discovery that two of these genes render an identical protein; and the discovery of four small introns in all the genes at this locus, among others. All these discoveries about the structure and function of my gene almost completed its molecular biology; the missing piece was its regulation [3-7].
Learning gene regulation was the next station in my journey and the best laboratory in the world to acquire such knowledge was in Strasbourg, France. The viral genome of the SV40 virus was the eukaryotic gene model with which I initiated my postdoctoral research on gene regulation in that Strasbourg´s laboratory. Its genome is very small [5,224 nucleotides or 5.2 thousand nucleotides or bases (kb)] compared with our genome (3 million kb), but it has the virtue of using the transcription apparatus of mammalian cells to operate [8]. The TATA box dictating where a gene´s transcription is initiated and the enhancer dramatically increasing this process, were among the world-renowned contributions of the laboratory I landed in [9]. I was given the task of discovering what was driving the high efficiency of the transcription of the early expressed genes of this viral genome. While others were using site-directed mutagenesis to characterize the virus enhancer, we used it to mutagenize each of the six GC-rich boxes distributed in pairs in three 21-b repeats [10]. We even altered the sequence of the pair of boxes within each of the three repeats. Each plasmid construct with the six individually mutated boxes and the three clones corresponding to the boxes mutated in pairs, were transfected into a cell line derived from kidney cells from a green monkey. With a so-called nuclease S1 protection assay, we quantified the level of expression originated from the early genes present along with their promoter in the transfected expression plasmids.
Through the analyses of the results we revealed for the world the nature of what is now referred to as the proximal gene promoter: in the SV40 genome it is the GC-rich boxes located precisely between the TATA and the enhancer; this is the answer [11]. This wonderful eukaryotic molecular biology model, with just two sets of genes (early and late expressed) and three major regulatory elements (TATA box, enhancer, and GC-rich boxes), was one of the simplest surrogate research models we could turn to for revealing the secrets of gene expression in higher organisms. Armed with the gene regulation tools acquired in France, I returned to Mexico and to my human genome model; the hGH locus.
On my return to Mexico, the GH locus in anthropoids provided me, with a far-reaching model to investigate gene regulation in the genome of higher organisms. In the case of humans, we had discovered its chromosomal location, described its composition as a pentagenic gene family, and determined the tissue and level of expression of its gene members [12]. Besides the emblematic gene named hGH-N, which is responsible for the production of HGH in the somatotrophs of the hypophysis, this locus contains an additional GH-type gene, hGH-V, which in the placenta produces a variant of the hypophyseal hormone that is postulated to replace it exclusively during the gestation period of pregnant women. It also includes the two above mentioned placental genes rendering an identical HPL, known as hPL-1 or hCSH-A and hPL-2 or hCSH-B, and finally, a gene referred as hPL-L or hCSH-L, which we found was virtually inactive, thus considered it a pseudogene [7,13].
Yet how was it that being encoded by genes showing more than 90% similarity in their flanking sequences (and even more in its coding ones), being so close to each other in the genome, and having been derived from a common ancestor not long ago, they display marked differences in their pattern of expression both temporal and tissue-specific? Not only is hGH-N set apart by its hypophysis-specific expression, but also by the fact its expression is shut down in pregnant women to allow the placental members of the family to control maternal metabolism to ensure the survival of the human fetus. Likewise, not only is the expression of the so-called placental genes is almost exclusive to the placenta, but this expression is also synchronized with the development of the placenta itself.
The coding sequences for these 5 genes amount to no more than 5kb, just as in the case of SV40. So, where do the secrets for regulation of its tissue-specific and temporal gene expression reside?
When in addition to the five coding sequences of the genes in the hGH locus, we consider their introns and flanking sequences, as well as the space separating them (intergenic regions), it turns out that this gene family encompasses ten times more genetic material than SV40 (approximately 50 kb). It was precisely by dissecting these gene-flanking DNA zones that we and other researchers were able to discover the main mechanisms regulating its differential tissue-specific expression [14]. One such mechanism is exemplified by Pit-1 (pituitary transcription factor 1, also referred as growth hormone factor-1) acting on the promoter of hGH-N to induce the transcription of this gene in the pituitary but unable to do so on the rest of the genes (GH-V and CSHs) as a result of their promoters containing a region of 0.26 kb, referred to as element P, to which certain placental proteins bind, interfering with Pit-1’s binding [12,15]. Another mechanism is the enhancer, which, as exemplified in SV40, is an expression control element that potentiates transcription of adjacent gene(s) [9]. In the case of the hGH locus it happens to be located only after the CSH genes, thus explaining the amazingly high production of CSH by the human placenta at the end of pregnancy, with quantities surpassing one gram per day [6]. Therefore, to control the tissular and temporal expression of a gene family, the human genome devotes ten times more DNA than that needed for specifying the hormones codified in this family.
My laboratory in Mexico joined the USA team exploring the unknown DNA regions surrounding the hGH locus in the five genes, beyond their proximal promoters and terminators [16]. Since it was our side project, it took us several years to achieve this monumental work that set a world-record then [17]. With today´s technology it could be redone in hours from start to end. But what did our quixotic achievement teach us about the use of sequencing the whole genome which took place almost fifteen years later? Well, that when you have the full sequence of your research model, many interesting projects arise from “mining” this genomic information.
In our case, wishing to prove the usefulness of having sequenced our tiny piece (0.0016%) of the human genome, we achieved the following inventions and discoveries: First, seeking to explain why some children with short stature fail to respond to biosynthetic or recombinant(r) GH, we designed a Polymerase Chain Reaction assay to diagnose gene deletions in the hGH locus; and thus, without meaning to do so, we invented what to the best of our knowledge, was the first companion diagnostic for a biological drug (rHGH)1. This genomic information-based diagnostic test not only served as the companion diagnostics for this inaugural product of the modern biotechnology, but also as a versatile tool to investigate gene alterations in the locus soon to be proven when explaining the extremely rare condition of absence of HPL in an otherwise uneventful pregnancy [18,19].
Second, we used the transcriptional unit of the hGH-N gene to reprogram yeasts to produce HGH that was conveniently secreted in the culture media in a correctly folded and thus active state [20]. Then we succeeded in doing so for the rest of the hormones coded in the hGH locus and a dozen plus animal GHs, CSHs and even prolactins, thus resulting in the world´s largest collection of recombinant GHs [21].
Third, we exploited the gene´s tissue specificity to develop viral vectors capable of selectively replicating within cell-cultured hypophyseal tumor cells that were then burst by induction of cell lysis, while leaving any other type of normal cells intact [22]. This experience further led to the development of a similar treatment but for cervical cancer successfully tested in an animal model and to the first clinical trial ever carried out in Latin America [23,24].
And fourth, we used this fabulous experience and confidence in our capabilities to launch a genomic biotech start-up that develops and offers innovative gene tests to aid in the diagnosis, prognosis, prediction, and even selection of the right treatment for disease, as well as, genetic and genomic manipulations as the basis for innovative solutions to the problems being faced by the biotech and food industries in our country [25,26].
1A FDA report refers to the approval of Herceptin and its companion test HerceptTest in September 1998, as the first major landmark in personalized medicine (www.fda.gov/downloads/scienceresearch/specialtopics/personalizedmedicine/ucm372421.pdf). However, our use of the sequence of the hGH locus to develop a genetic test usable to predict response to rHGH, published as a BSc thesis in 1994 and as a scientific paper in 1997 (18) is, to our knowledge, the first such achievement and thus a pioneer contribution to the era of personalized medicine.
With support from Genome Quebec, our sequencing of the GH loci in bacterial artificial chromosomes of species from all the different groups of primates enabled us to determine the composition and organization of their GH locus. Our findings confirmed a single GH-type gene in prosimians, more than half a dozen GH-type genes (including pseudogenes) in new world monkeys (NWM), two GH-type genes mixed with four CSH-type genes in old world monkeys (OWM), and between four and six GH/CSH genes in great apes [27].
This dramatic evolutionary story illustrates two opposite tendencies. In the beginning, at the transition from prosimians to NWM the locus quickly went from a single GH-type gene to, in some cases, dozens of copies of it, some being active and some harboring structural alterations known to render pseudogenes [28]. Then, at the transition from NWM to OWM, the tendency was to keep gene count around half a dozen, at the same time that some of the GH-type copies differentiated to become CSH-type genes [29]. Also, pseudogenes were not especially common in the new loci.
On top of these evolutionary enigmas, is the story of great apes (gorilla/chimpanzee/human). We found a clear tendency towards reduced numbers of different CSHs, without necessarily reducing the gene count, from three in the gorilla, passing through two in the chimpanzee, to only one in humans [7,30,31]. In turn, intergenic regions expanded and differentiated to control differential expression of the new members in the placenta. In particular, the above referred enhancers were acquired from an adjacent locus early in evolution and when the gene duplication-expansion occurred, ended up after the CSH genes only [27].
To investigate the genomic underpinnings of one of the most feared tumors in Mexico, cancer of the uterine cervix, we teamed with researchers at the Broad Institute in Boston, at the Mexican National Institute of Genomic Medicine in Mexico City, and at other international leading Universities and Institutes, to perform whole exome sequencing (only protein-coding regions of the genome) on samples from 115 cervical cancer patients from Norway and Mexico. When compared with reading the hGH locus, this task represented close to a hundred thousand-fold increase in nucleotide reading. This huge amount of genomic data derived from cervical cancers was compared with that of normal control tissue.
In summary, this study demonstrated relationships between recurrent somatic mutations, copy-number alterations, gene expression, and human papilloma virus (HPV) integration in cervical carcinomas. We found significantly recurrent somatic mutations in the MAPK1 gene in squamous cell cervical cancers, to our knowledge the first such report in human cancers. In addition, we revealed evidence of potential ERBB2 activation by somatic mutation, amplification, and HPV integration, suggesting that some cervical carcinoma patients could potentially be considered as candidates for clinical trials of ERBB2 inhibitors. Furthermore, our data suggested that alterations in immune response genes may synergize with HPV infection in the pathogenesis of squamous cell carcinomas. Finally, our data also suggest that the association between HPV integration and increased expression of adjacent genes is a widespread phenomenon in primary cervical carcinomas [32].
In this way, we revealed the landscape of genomic alterations associated with the genesis of cancer of the uterine cervix.
The reading of the genomes of cohorts by NGS leading to the discovery of these genes illustrates not only the benefit of genomic research on disease diagnosis but also offers the possibility of using them as targets for new drug development.
World-class medical research increasingly relies on tools from the “omics” disciplines (genomics, epigenomics, transcriptomics, and proteomics, mainly). Providing quality biospecimens for these studies, collected and processed with special care to preserve as much as possible their intactness and thus richness of information, is the role of biobanks. These biorepositories or biobanks support the transportation, storage, preservation, and initial histopathologic and analytical examinations of bio specimens; as well as the procurement of all relevant clinical information. By doing so, biobanks act as essential bridges and effective catalysts for research synergies between basic and clinical sciences [33].
At our institutions, with support from the Mexican Council of Science and Technology, we have been migrating our efforts of simply collecting biospecimens to satisfy our urgent and short-term needs of experimental materials required for our students´ theses and the grants of our colleagues, to adopting good bio banking practices throughout the process [34]. By realizing that this limited exploitation of such valuable resource is a common problem among basic and translational research biomedical laboratories in our country - as it is in most other countries too-, this effort is now leading to the creation of a national network of biobanks, which we believe will improve the competitiveness of our national medical research, and better serve its clinical, pharmaceutical and biotech sectors. Leading biobanks around the world are supporting the sequencing of the entire genomes of thousand if not million of patients participating in complex diseases research projects and in clinical trials for new drugs, a task whose genomic scale is in the order of trillions (3x109 x 1,000 patient cohort or 3x1012, or x 1,000 000 soon to be reached-population size genomic study or 3x1015).
With advances in technology, the unravelling of secrets concealed in our hereditary encyclopedia has called for ever-increasing readings of our genetic code. In this story of my journey from genes to genomes the amount of information to deal with grew exponentially. Computers and software packages once convenient but not essential tools became indispensable with the switch from genetic to genomic research. Likewise, biobanks, which now make it possible for scientists around the globe to share valuable samples treasured in their cohorts. Combined with the eras of digital health and population genomics (already surpassing a million individual genomes sequenced they will be the pillars supporting the massive genomic information highways already under construction for our youth to undertake their own journeys [35].
As the different approaches during my journey show and Figure 1 summarizes, my destination in this adventure has been the starting point for the new hunters of these much-pursued secrets.
The author declares that there is no conflict of interest regarding the publication of this review paper.
The author financed this research odyssey by successfully competing for funds from CONACyT, UANL, Fondo Ricardo J. Zevada, FUNSALUD, the Mexican Ministries of Education and Economy, and UNESCO, mainly; also, through donations from General Foods, IBM, CEMEX, and CyDSA, among others.
The author wishes to dedicate this review to the outstanding teachers and mentors who where his most valuable source of inspiration; especially to the late Grady F. Saunders, Francisco J Sánchez-Anzaldo, and Pierre Chambon.