A Possible Interaction or A Similarity between the Mechanism of Action of the Arylamidase Enzyme and the Ferripyoverdine Receptor Genes in Pseudomonas aeruginosa
The essential element iron is relatively soluble in nature and because of its relative solubility, gram negative bacteria, for example, Pseudomonas aeruginosa synthesizes a low molecular weight compound called Pyoverdine. Pyoverdine enables P. aeruginosa trap iron from the external environment into the internal milieu. Iron bound to these compounds are recognized at the outer membrane by the ferripyoverdine receptor genes. The ferripyoverdine receptors help in channeling iron into the internal milieu for use by P. aeruginosa. The Pseudomonas aeruginosa arylamidase enzyme was purified in 1971, and P. aeruginosa uses this enzyme to hydrolyze basic and neutral N-terminal amino acid residues from amino-ß-naphthylamides, dipeptide-ß- naphthylamides, and a variety of polypeptides and according to the scientists who purified this enzyme, only those substrates having an L-amino acid with an unsubstituted α-amino group as the N-terminal residue were susceptible to enzymatic hydrolysis. Based on scientific observations from my previous work titled “Ferripyoverdine receptors and general metabolism in Pseudomonas aeruginosa, Preliminary results” and the scientific research work done by other authors, I am of the opinion that the ferripyoverdine receptor gene and the arylamidase enzyme may interact or may be somewhat similar in their mechanisms of operation.
Keywords: Chloramphenicol; Pyoverdine; Iron; Amino acid; Inducer; Arylamidase Enzyme; Ferripyoverdine Receptors; Pseudomonas aeruginosa; Media Composition; Gene Expression
Pseudomonas aeruginosa is a gram negative bacterium widely distributed in the natural environment. It was first isolated in 1872 by Schroeter and later identified from green pus by Gessard in 1882 [1]. It can thrive in any environment because of its high adaptable nature and apart from the soil being its primary habitat, it can also be found in hot zones and in environments with limited nutrition. It is a major pathogen affecting plants, animals and humans. Clinically, P. aeruginosa plays an important role in the survival of patients having infections that has been colonized by it [2].
Based on scientific observations from my previous work titled “Ferripyoverdine receptors and general metabolism in Pseudomonas aeruginosa, Preliminary results” [3] and the scientific research work done by Bishop et al., 2017 and Riley and Behal, 1971, a possible interaction or similarity in the mechanisms of operation of the ferripyoverdine receptor gene (P. aeruginosa ) and the arylamidase enzyme is suggested in this article which would be explained further under various subheadings:
Iron is an important metal and its importance has been extensively studied in different organisms [4, 5]. It has been studied to play a crucial role in the expression of some enzyme of the citric acid cycle, and apart from being important, it is relatively scarce and insoluble [6-9]. Fluorescent pseudomonads therefore produce pyoverdine to enable iron acquisition during the periods of insolubility and scarcity [10].
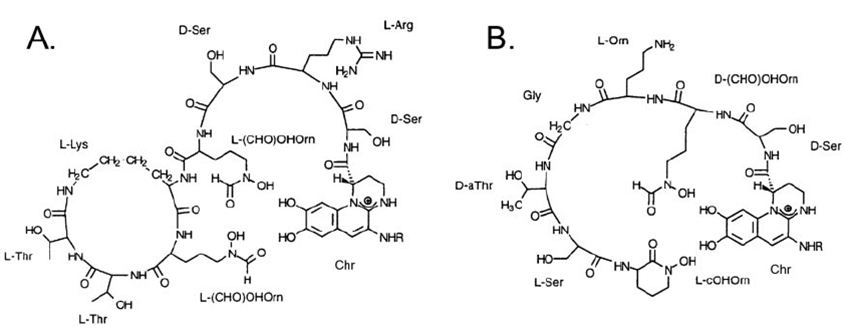
Pseudomonas aeruginosa synthesizes pyoverdine when iron becomes insoluble and scarce. Structurally, pyoverdine consists of an invariant core ( the fluorescent chromophore derived from 2,3-diamino-6,7-dihydroxyquinoline ) that is modified by the addition of about 6-14 amino acid chains depending on the producing strain and strains of P. aeruginosa secrete one of three pyoverdines. The structures of types I and II is as shown below[Figure 1];
Pyoverdine is synthesized under iron limiting conditions, and exposure to the metal cadmium was previously demonstrated to contribute to pyoverdine abundance in P. aeruginosa [13,14]. About 12 proteins take part in pyoverdine biosynthesis which has been implicated to take place, starting from the cytoplasm, and ending in the periplasm [15-19].
P. aeruginosa has been studied to be in possession of a complex genome and its genome is found to contain about 19 extracytoplasmic function sigma factors (ECF). Extracytoplasmic function sigma factors are found in many bacteria and they are small regulatory proteins which direct gene regulation in response to stress or a number of environmental stimuli [20-30]. In P. aeruginosa, the extracytoplasmic function sigma factors (fpvI and pvdS ) are responsible for the transcription of the pyoverdine biosynthetic genes and the ferripyoverdine receptor gene, the P. aeruginosa . Also present is the fpvR, a presumed cytoplasmic membrane-associated anti-sigma factor controlling the activities of these extracytoplasmic function sigma factors. The fpvR binds to fpvI and pvdS, inhibiting their activities by preventing their interaction with core RNA polymerase.
Pyoverdine biosynthesis is followed by a subsequent expression of the receptor gene called the P. aeruginosa which aids in its internalization. Upon internalization by P. aeruginosa , pyoverdine has been studied to be translocated from the periplasmic space to the extracellular milieu [31]. The ferripyoverdine receptor of P. aeruginosa (P. aeruginosa ) was cloned and identified to have a molecular mass of 89,395 daltons [32,33]. In addition to playing the role of a receptor to iron bound to pyoverdine, the P. aeruginosa gene has also been hypothesized to be targeted by antibiotics and bacteriophages and serve as entry points for them and during my PhD thesis, I could successfully identify variants (apart from the already existing P. aeruginosa genes) of the ferripyoverdine receptor genes, and furthermore, scientists have previously discussed the clinical importance of this gene in P. aeruginosa and the possibility of harnessing this entry point as an avenue for drug discovery has been proposed [5,13,34, 35, 37, 39].
Pseudomonas aeruginosa uses an alternate receptor called the fpvB (apart from the P. aeruginosa ) to internalize iron bound to pyoverdine. The fpvB was identified following the observation that an P. aeruginosa mutant could still grow in the presence of its cognate pyoverdine under iron limitation [38]. Recently, a variant of the fpvB gene was identified by me and from evolutionary point of view, these genes maybe evolving to suit the need of Pseudomonas aeruginosa as regards iron internalization and usage [36].
The arylamidase enzymes are widespread in nature. They were initially thought to be characteristic of gram negative bacteria alone. However, other studies have shown them to be present in gram positive bacteria, animals and humans as well. Although various functions has been suggested for this enzyme other than protein synthesis for cellular multiplication, in the rat for instance, this kidney enzyme functions as an angiotensinase or an amino polypeptidase and their role in protein and peptide metabolism in the nervous system has been determined [40,41]. The intracellular and constitutive arylamidase enzyme of Pseudomonas aeruginosa was purified decades ago by several processes which included salt fractionation, ion exchange chromatography, gel filtration, and lastly adsorption chromatography. This enzyme was estimated to weigh about 71 kDa and P. aeruginosa uses this enzyme to hydrolyze basic and neutral N-terminal amino acids from amino-ß- naphtylamide , dipeptide-ß-naphtylamide and a variety of polypeptides [42].
In the past, media composition has been shown to play an important role in the expression of some genes [3-47]. In iron rich media for example, the receptor genes for iron bound to pyoverdine (ferripyoverdine complex) are not expressed. The P. aeruginosa gene is expressed in a deferrated medium containing glycerol as carbon source while the fpvB gene is induced under iron limitation, in Casamino acid (CAA) and M9 –glucose media [38]. Subsequently, the arylamidase enzyme activity was observed in one media not containing iron (TYE) and not in another containing iron (GBS).
The aim of this article is to further highlight the importance of media composition on gene expression and the probable role the ferripyoverdine receptor genes might play in relation to general metabolism in P. aeruginosa and because of the similarities which may probably exists between the activities of the ferripyoverdine receptor gene (P. aeruginosa ) and arylamidase enzyme of P. aeruginosa, further research work is proposed. The outcomes of these future scientific experiments, if carried out, may possibly help in increasing the already vast available knowledge on the ferripyoverdine receptor genes and also provide more information about the arylamidase enzyme and its mode of operation.
As previously described by Osayande, 2013. Briefly, I used P. aeruginosa wild type (PAO1) and ferripyoverdine receptor mutants PAO1-pvdD pchEF FpvA, PAO1-pvdD pchEF FpvB, and PAO1-pvdD pchEF FpvAFpvB (as previously described under the subheading: strains used in this study by Osayande, 2013, and also by Ghysels et al., 2004) as inoculums for the VITEK 2 biochemical instrument for bacterial identification. The inoculums were prepared as suspensions from cotton swab of sample bacteria (P. aeruginosa wild type and ferripyoverdine receptor mutants) applied directly to 3ml of sterile water or sterile water of same volume containing either 100μM iron or 100μg/ml gentamicin or both, adjusted to a turbidity of about 0.55. The VITEK 2 instrument automatically filled, sealed, and incubated the individual test cards with the prepared culture suspension. The concentration of gentamicin (100μg/ml, Ghysels et al., 2004) was also used in the reaction mixture for wild type (PAO1) and ferripyoverdine receptor double mutant (PAO1-pvdD pchEF FpvAFpvB) [38]. Optical readings were taken automatically and based on these readings; an identification profile was established and interpreted according to a specific algorithm. Results obtained were compared to the database, generating an identification listed as “excellent,” “very good,” “good,” “acceptable,” or “low discrimination” (which are considered correct) for an unknown organism [3].
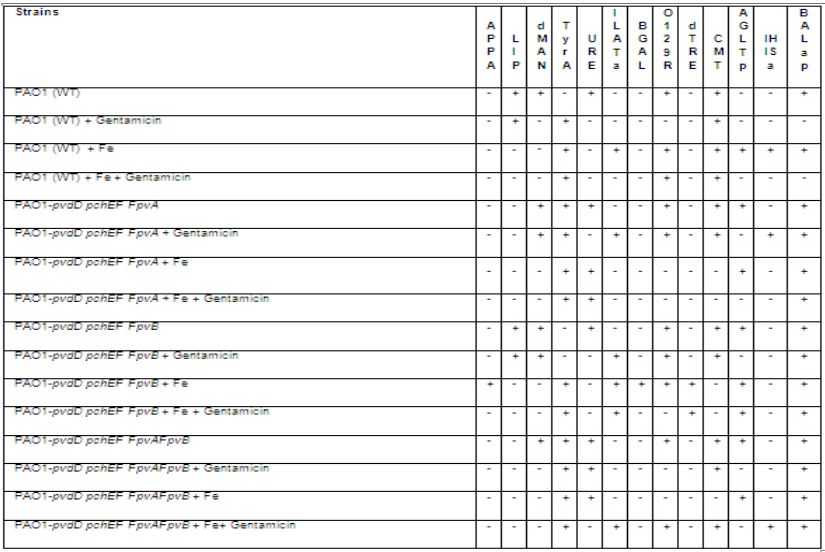
The identification of Pseudomonas aeruginosa wild type and mutant strains used is as shown in the Table 1 below:
Surprisingly, the results of bacterial (Pseudomonas aeruginosa wild type and mutants) identification from the VITEK machine were readable. While the wild type was identified as 98% “Pseudomonas aeruginosa”, the mutants were scored less (97%). And not only was P. aeruginosa wild type and mutants identified, its ability to utilize the different substrates impregnated in the VITEK cards were also recorded by the machine. Using about 100μM of iron, and 100μg/ml gentamicin, the gentamicin resistant allelic mutant of P. aeruginosa (PAO1-pvdDpchEfFpvB; a mutant with an operational P. aeruginosa gene) tested positive to the APPA (Ala-Phe- Pro-Arylamidase) enzyme activity (See Table 2 attached here and also Osayande, 2013). The amount of iron I used was 100μM, this amount is very minimal compared to the amount of iron present in the GBS medium used by Riley and Behal (1971), and the observable enzyme activity recorded by the VITEK machine may not have been prevented by this amount of iron. Questions which may arise from these experiments are those asking how much iron is needed to be present in a reaction mixture in order for it to completely negate gene expression [49].
Also impregnated in the VITEK 2 cards are TyrA (Tyrosine arylamidase) , AGLTp (Glutamyl Arylamidase), BALap (Beta- alanine arylamidase). Although, more substrate hydrolysis was observed for these other amino acid - arylamidases than for the APPA, the reason behind the differences in the observed hydrolysis is unknown at this moment. Interestingly, some amino acids are more soluble than others and they vary in nature. This variation (structural or physical) may contribute to the level of activity of the different amino acid-arylamidases incorporated into the VITEK cards, more investigations are also required in this regard.
Decades ago, precisely in 1971, the P. aeruginosa arylamidase enzyme activity was observed in the TYE and GBS media ( Table 3). The ability of cells grown in these two media to hydrolyze the substrate “Amino Acid – β-Naphtylamide” was studied.
Intact GBS-grown cells lacked the ability to hydrolyze the substrate amino acid- ß –naphtylamide whereas intact TYE-grown cells completely hydrolyzed the substrate. It will be interesting to note that in this experimental system, the hydrolysis or uptake of the substrate was induced by amino acid and not by amino acid - β-naphtylamide itself which typically is the substrate.
Furthermore, pre-incubation of the GBS-grown cells with an antibiotic (chloramphenicol) before the addition of an inducer (the inducer here is an amino acid) completely inhibits the response of these cells to the inducer. In addition, when the inducer was added, a period of no activity was observed. This observable period of delay, was subsequently followed by substrate uptake [40].
According to Riley and Behal, the period of no activity or delay could have meant that some proteins needed to be synthesized before the GBS-grown cells could completely hydrolyze or assimilate its substrate.
In their experiment, Bishop et al., (2017) recently demonstrated that the transport of ferripyoverdine complex by the ferripyoverdine receptor gene (P. aeruginosa ) present on the outer-membrane of P. aeruginosa causes the proteolytic degradation of the anti-sigma factor they tagged fpvR20, inducing the expression of the sigma factors pvdSand fpvI , following their release from the anti-sigma factor. In addition, a delay is observed when the sigma factor fpvI released from the anti-sigma factor fpvR20 is supplemented by newly synthesized fpvI .
Chloramphenicol is a translational inhibitor and its inability to prevent the induction of the expression of the P. aeruginosa gene was recently demonstrated by Bishop et al., 2017 [50-52].
When Bishop et al., added chloramphenicol to their reaction mixture during pyoverdine –mediated induction of gene expression, a slight delay in the induction process (here, pyoverdine is used as the inducer) was observed and according to them, the supplementation of the previously existing fpvI with the newly synthesized one could provide an explanation for the observed delay in the induction of the expression of the P. aeruginosa gene during pyoverdine -mediated induction of gene expression [50].
During amino acid -mediated induction of arylamidase activity in GBS-grown cells preincubated with chloramphenicol, the observed delay was meant to be accounted for by the “probable synthesis of some proteins” whereas in the experiment carried out by Bishop et al. 2017, the observed period of delay in the presence of chloramphenicol and the inducer could be due to the “supplementation of an existing ECF (fpvI ) with a newly synthesized one”. I am of the opinion that these two gene operational systems may have something in common based on these different observations.
From the work done by Riley and Behal, (1971) and Bishop et al. (2017), the probable observable similarities made me ask these questions below that may only be answered in future scientific experiment (s). These questions, amongst others, are:
• Why was the arylamidase enzyme inactive in one medium (GBS) and active in another medium (TYE), was the enzyme activity
observed in the TYE medium as a result of the absence of iron in it and is the arylamidase activity regulated by iron? [53]
• According to Riley and Behal, some proteins were needed to be synthesized before the GBS-grown cells could completely
hydrolyze its substrate. What proteins were needed to be synthesized? (As experienced by Bishop et al. 2017)
• Following the work of Bishop et al. (2017) and Riley and Behal, (1971), could the arylamidase enzyme and the ferripyoverdine
receptor gene is similar in their manner of functioning? (My opinion).
• The GBS medium contained about 0.1g of Ferrous chloride, was this amount of iron enough to repress the arylamidase enzyme
activity or something else was responsible for enzyme inactivity? [49]
• If it is true that arylamidase enzyme activity is iron repressed, then, the next question would be to ask if chloramphenicol was
able to relieve iron-repression of the arylamidase enzyme activity or whatever gene (s) present at the time of reaction as observed
by Bishop et al. 2017 of the P. aeruginosa gene and chloramphenicol during pyoverdine –mediated induction of gene expression.
• During the work of Riley and Behal in 1971, was the P. aeruginosa receptor gene also expressed in the reaction medium? (My speculation).
• If yes, was the chloramphenicol used by Riley and Behal in 1971 affecting the arylamidase enzyme activity or the P. aeruginosa gene
expression or both? [50]
This suggestion is made as a result of the fact that only the gentamicin resistant allelic mutant of P.aeruginosa (PAO1-
pvdDpchEfFpvB; a mutant with an operational P. aeruginosa gene) tested positive to this enzyme activity (
Table 2).
• Does the P. aeruginosa gene contribute to the activity of the arylamidase enzyme or their activities are intertwined or they simply have
the same mode of operation?
Although, the ability of the other elements present in the GBS medium to influence the arylamidase enzyme activity should not be underestimated, the clear picture of what might have occurred decades ago may probably be traced to the presence of iron in one medium (GBS) and its absence in the other medium (TYE) or reasons yet to be identified or speculated.
A quick Summary of the “Possible” similarities between arylamidase enzyme and ferripyoverdine receptor gene is further explained in the Table 4 below:
The results obtained from my work titled “ Ferripyoverdine receptors and general metabolism in Pseudomonas aeruginosa , Preliminary results” during my PhD thesis, the scientific research work done on the arylamidase enzyme of P. aeruginosa and the recent scientific observations made by Bishop et al. 2017, and those of other authors can be used to re-study the arylamidase enzyme activity in Pseudomonas aeruginosa and it may be interesting if its activity can be linked to the ferripyoverdine receptor genes [3,42,50].
A detailed future experimental research explaining what happens when metals, for example, iron, and in addition, when antibiotics like Chloramphenicol and Gentamicin, are added to a minimal or complex media for bacterial growth (Pseudomonas aeruginosa) is suggested in order to fully understand how genes, especially those of pathogenic organisms are expressed in relation to the presence of these aforementioned substances in a reaction mixture. Such detailed experiments may help us understand that the ferripyoverdine receptors are not only expressed in relation to iron unavailability or scarcity or other circumstances yet to be identified, they may also contribute to the general metabolism in P.aeruginosa. This additional knowledge may serve as an alternative avenue for drug discovery against highly pathogenic organisms like P.aeruginosa and the results presented here can be used freely by any laboratory for further investigations. Hopefully in the near future, more answers will be provided to unravel the hidden information, if any, between these two systems (The ferripyoverdine receptor genes and the arylamidase enzyme) of Pseudomonas aeruginosa.
This work was done through the support of the VUB doctoraatsbeurs and the author is grateful to the Vrije Universiteit Brussels (former employer) for the provision of academic scholarship.