Association of Begomovirus with Okra (Abelmoschus esculentus L.) leaf curl virus disease in southern India
A severe leaf curl disease of Okra with high disease incidence and loss of production was observed during field survey of various locations of Southern India, and causal organism was identified as Begomovirus and its association was confirmed by polymerase chain reaction (PCR) but also by cloning and sequencing of amplified PCR products and whitefly transmission. The DNA β and Coat protein gene was cloned and sequenced. The sequence data were assembled and analyzed by using the Bio-Edit (version 5.0.9). The nucleotide sequences of DNA β and Coat protein gene showed highest identities with Bhendi yellow vein mosaic virus isolates. But the most important and novel observation was recorded that the infected plants with severe leaf curl disease never develops yellow vein mosaic symptoms during same field conditions. These findings suggest that the association of Begomovirus with severe leaf curl disease could be strain of Bhendi yellow vein mosaic virus occurring in southern India. The above findings require more study to confirm strong association of causal organism with this unique symptom.
Keywords: COkra (Abelmoschus esculentus L.); Leaf curl disease; Begomovirus; DNA β; Polymerase chain reaction; Cloning
Okra (Abelmoschus esculentus L.) is an important vegetable crop of India cultivated in 452 hectares and yielding 4803 tones [1]. Okra leaf curl disease is an emerging serious disease in India. Severe leaf curling symptoms were observed while at the same time some plants showed only yellow vein mosaic in the same field. Severe leaf curling with vein thickening symptom of okra have been observed over the years and posed a serious threat to the Okra cultivation in India. Bhendi yellow vein mosaic virus disease has been reported earlier in India [2-4]. Recently, an association of bipartite begomovirus with Bhendi yellow vein mosaic disease in India has also been reported [5]. Naturally infected plants of Okra develop symptoms like severe leaf curl, vein thickening with stunted plant growth (Figure 1). Recently, associations of betasatellites and alphasatellites causing leaf curling disease in okra have been reported from India [6,7] and southwestern Cameroon [8]. Recently, 36 different betasatellites isolated from okra exhibiting severe leaf curl and yellow vein mosaic symptoms has been determined from northern India and sequences segregated into four groups, two groups corresponds to betasatellites that have been previously identified in okra from the sub-continent. Two new group of betasatellite was identified as Bhendi yellow vein India betasatellite (BYVIB) and Croton yellow vein mosaic betasatellite (CroYVMB) in okra from northern India [6]. Recently, an association of alphasatellite DNA with Okra enation leaf curl virus (OELCuV) causing enation and leaf curling in okra (Abelmoschus esculentus) plants has been reported for the first time from India. The full-length DNA contains 1350 nucleotides which showed typical genome organization of an alphasatellite. The highest nucleotide sequence identity (79.7%) was found with Hollyhock yellow vein virus-associated symptomless alphasatellite (HoYVSLA) [7].
The naturally infected okra leaf sample was used for virus acquisition. The healthy whiteflies were allowed to feed on infected leaf for 24 hours and then subsequently those viruliferous white flies were feed in healthy okra seedlings under insect proof microcages for 24 hrs. The inoculated seedlings were kept inside the insect proof green house for symptoms development and the inoculated plants developed same symptoms as in the field. To identify the Begomovirus infection, total genomic DNA was isolated from both naturally as well as artificially infected Okra leaves and polymerase chain reaction (PCR) was performed using primers sets specific to DNA β F 5’-GGT ACC ACT ACG CTA CGC AGC AGC C-3’ and DNA β R 5’-GGT ACC TAC CCT CCC AGG GGT ACA C-3’ and Coat protein gene CPF5’-TTA TGT CGA AGC GAG CTG CC-3’ and CPR5’-TTT CAA TTC GTT ACA GAG TCA TA-3’ [2,3] resulted in amplicons of 1.4 kb and 750 bp DNA fragments respectively. PCR reactions were performed in 50 μl total volume using 100 ng template DNA, 2.5 units of Taq DNA polymerase (MBI Fermentas, USA), 1x PCR buffer and 0.2 mM dNTps, 50pM of each primer. The PCR was carried out in a Eppendorf master cycler (Gradient S) at 94 0C for 5 min (pre-heating), followed by 35 cycles at 94 0C (denaturation) for 1 min, 56 0C (annealing) for 1 min, 72 0C (extension) for 1 min, and 72 0C (final extension) for 7 min for DNA-β gene detection and 94 0C for 2 min (pre-heating), followed by 35 cycles at 94 0C (denaturation) for 1 min, 55 0C (annealing) for 1 min, 72 0C (extension) for 1 min, and finally 72 0C (final extension) for 5 min for coat protein gene detection.
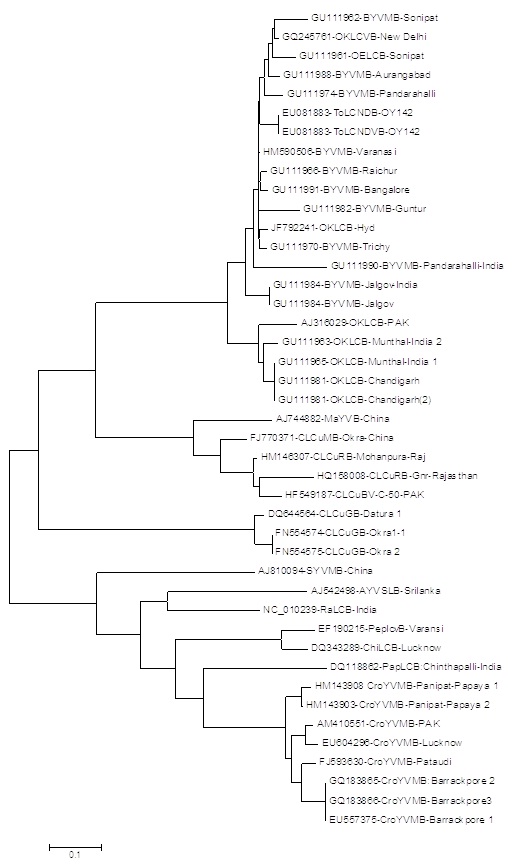
The PCR amplified products from Hyderabad isolate was cloned and sequenced and submitted to GenBank. The coat protein sequences consisted of 771 nucleotides (Accession Number JF792242) whereas DNA β sequence consisted of 1355 nucleotides (JF792241). The complete nucleotide sequence of coat protein gene and DNA β were initially searched for similarity using BLAST programme (http://www.ncbi.nlm.nih.gov/BLAST) [9] and the sequence which showed higher score were selected for further analysis. The sequence was analyzed to determine percent identity matrix with those species showed maximum identity during BLAST search. The nucleotide sequence of the complete DNA β shared highest (92.9%) and lowest (39.1%) identities with the isolates of Bhendi yellow vein mosaic virus (BYVMB-Trichy-GU111970), and (BYVMB-Raichur-GU111966) and Croton yellow vein mosaic betasatellies-Lucknow (CroYVMB-Lucknow-EU604296-).The nucleotide sequences of coat protein gene showed highest (99%) similarities with the various isolates of BYVMV (AJ278861, AF465619, FN645917) and shared lowest (92%) identity with the isolates of BYVMV (FJ561298) and MYMBV (EU360303). The phylogenetic tree was constructed by using beta DNA sequences of selected isolates and the closest homology was observed with BYVM betasatellite [India: Trichy:OY118:2006] followed by other okra leaf curl virus (Figure 2). On the basis of above information and result obtained, it can be concluded that the Okra leaf curl virus disease observed in Hyderabad and Guntur, India is caused by Begomovirus associated with DNA β and this requires some deeper study to strongly confirm by Agroinoculation.
Authors extend their thanks to Mr. SK Gupta (President, JK Agri Genetics Ltd. Hyderabad, India) for providing necessary facilities, guidance, valuable suggestions, and continuous encouragement.