DNA Sequencing of Chilling Tolerant Phaseolus Vulgaris L. and Chilling Sensitive Vigna Unguiculata L.
The response of dehydrin gene in P. vulgaris L. and V. unguiculata L. to LNT (Low night temperature) stress was studied. It was inferred that dehydrin gene is induced to express the protein which confers an adaptation effect only in P.vulgaris L. but not in V. unguiculata L. LNT did not show any genetic variations when compared with a non LNT stress induced plant. Hence P. vulgaris L. is considered to be cold tolerant plant and V. unguiculata L. is a sensitive plant. P. vulgaris L. and V. unguiculata L. (LNT treated and control) can be analysed for other genes involved in cold stress. This observation hypothesizes that there is no significant gene level variations between LNT stress induced plant and control plants. It can also be hypothesized that variations may be occurring in its expression level to code for the protein responsible for making the plant tolerant to cold. Moreover the present study also suggested no significant gene level variations between P. vulgaris L. and V. unguiculata L. plants even though significant biochemical variations were observed when these plants were exposed to LNT stress.
Keywords: Dehydrin Gene; LNT Stress; Cold Tolerance; Gene Expression
Crop plants are exposed to a range of external abiotic stress factors such as drought, salinity, cold, freezing, high light sensitivity, nutrient imbalances etc, Numerous cold induced genes have been isolated and characterized in a number plant species.
An increasing number of genes have been revealed to be induced during cold acclimatization [1,2]. Many of these genes encode proteins with known freezing tolerance. They encode newly discovered proteins such as the Arabidopsis COR6.6, COR15a and COR78 polypeptides or homologous of LEA proteins such as Arabidopsis COR47. The polypeptides encoded by these cold-responsive genes fall into a number of groups based on amino acid sequence similarities, but all share the property of being extremely hydrophilic. In addition, many have relatively simple amino acid compositions (i.e. are composed largely of a few amino acids) have repeated amino acid sequence motifs and remain soluble upon boiling in dilute aqueous buffer. Among the highly expressed cold responsive genes of Arabidopsis are the COR genes also designated LTI (low temperature induced), KIN (cold-inducible) and RD (responsive to desiccation). The COR genes comprise four gene families, each of which is composed of two genes that are physically linked in tandem array. The COR78, COR15 and COR6.6 gene pairs encode newly discovered polypeptides and the COR47 gene pair encodes homologous of LEA group II proteins (also known as dehydrins and LEA D11 proteins). Recent studies indicate that COR15a acts in concert with other COR genes to enhance freezing tolerance [3,4].
LEA proteins are expressed at different stages of late embryogenesis in seed embryos and under various conditions of stress including desiccation. LEA proteins are found in different tissues and in all cell types. They have been found to accumulate in cytoplasm and plastids. The very nature of this wide cellular distribution infers a protective function [3]. Furthermore, LEA proteins and heat shock proteins have been shown to be involved in protecting macromolecules such as enzymes and lipids. Dehydrins (LEA D11 family) are proteins that occur in plants due to dehydration, low temperature, and osmotic stress. Earlier Inheritance studies including QTL analysis in crop plants revealed apparent co-segregation of Dhn genes with phenotypes associated with drought and freezing [5,6]. Dhns are unified by the presence of one or more copies of a putative amphipathic α-helix forming domain (The K-segments) which is highly conserved in higher plants. This and other distinct domains of Dhns including a phosphorylatable (the S-segment) and an N- terminal consensus sequence (the Y-segment) are pieced together in a consistent manner, interspersed by other lesser conserved and usually repeated domains (the F-segments) [7,8].
The assembly of domains into numerous, yet consistent permutation has resulted in a range of Dhn polypeptide lengths from 82 to 575 amino acid residues. The number of occurrences of the K-segment varies from one to 11 within a single polypeptide. The bulk of the Dhn polypeptide in most cases contains regions or domains (the F-segments) that are rich in Gly and polar amino acids (especially Thr) and are tandemly repeated between K-segments. But there are contrary examples were the F-segments located between the K-segments are rich in other amino acids or do not exit as tandem repeats. For example, the F-segments located between K- segments in all SK3 Dhns are not Gly rich, but in many cases are rich in Pro and Ala. Because of this distinction and the fact that the SK3 and some other Dhns tend to contain high percentage of acidic residues, it has been proposed that the Dhn family may contain biochemically distinct acidic sub groups. Two studies have clarified the location of Dhns in the cytoplasm [10]. Most Dhns contain putative bipartite nuclear targeting signal sequences. The predicted molecular weights of Dhns based on amino acid sequences are invariably less than their apparent molecular weight SDS-PAGE. This anomaly is also observed with Dhns translated in-vitro, retarded migration thus seems to be due at least in part to secondary structure in 0.1% SDS. A ~35 kDa Dhn of V. unguiculata L. which is associated with an increment of chilling tolerance during seedling [10]. A simple interpretation of these observation is that Dhns are lipid binding proteins possibly, Dhns and other LEA and COR (cold responsive) proteins function in a lipoprotein environment, at an interface between phospholipids bilayers and aqueous compartment. Dehydrins are a D11 family of late embryogenesis abundant proteins that are induced in vegetative tissues in response to low temperature. The main aim is to isolate the dehydrin gene from cold stress induced in-vitro cultured P. vulgaris L. and V. unguiculata L. plants and to analyse if any sequence variations occurs in the gene level.
P. vulgaris L. (Sel.9) and V. unguiculata L. (P.152) seedlings were grown under controlled climatic condition (in a polyhouse) by irradiance of 1500 μmol m-2S-1 (PAR), temperature of 28 °C and 70% RH for 7 days. The plants were watered regularly. The plants were classified into two groups according to the treatment. One group of plants was continuously grown under ambient irradiance and temperature controlled polyhouse, while the other group of plants were subjected to LNT (low night temperature) treatment of 15 °C by transferring the plants to chilling chamber and treatment was given for 12 h daily (each night from 18:00h to 06:00h) for 5 days. During daytime the plants were grown under polyhouse condition. On 13th day, total RNA was isolated.
100 mg of plant tissue was taken for the RNA isolation. The plant tissue was homogenized with 450 μl of RLT buffer and 150 μl of RLC buffer with the help of the homogenizer. The lysate was transferred into the QIA shredder spin column (lilac tube). The spin column was centrifuged at maximum speed for 3 min. After centrifugation, the supernatant of the flow through was transferred to the new microfuge tube without disturbing the cell debris. 225 μl of 100% ethanol was added into the sample and it was mixed immediately by pipetting. Then the sample was applied into the mini spin column and centrifuged for 1 min. at 10,000 rpm. The flow through was discarded and 700 μl of RW1 wash buffer was added to the column. The tube was centrifuged for 1 min. at 10,000 rpm. In the next step the flow through and the collection tube were discarded. The column was placed in a fresh collection tube and 500 μl of RPE was added into the column and centrifuged for 1 min. at 10,000 rpm to wash the column. The column was centrifuged again for 1 min. to dry the membrane to avoid ethanol. The flow through and the collection tube were discarded. Then the column was placed into a new 2 ml collection tube. Then 36 μl of RNase free water was added to the column to elute the RNA in a collection tube and centrifuged for 2 min. at 10,000 rpm. The isolated RNA was visualized in the 1.2% formaldehyde agarose gel electrophoresis. The isolated RNA was amplified by RT-PCR. 12 μl of amplified PCR product was loaded in the 1% agarose gelelectrophoresis [11,12].
5 volumes of PB buffer and 1 volume of PCR sample were taken in Qiaquick kit tube. Qiaquick spin column was placed in a provided 2ml collection tube. To bind DNA, the sample was applied to the Qiaquick column and centrifuged for 30-60 seconds. The flow through was discarded and the Qiaquick column back was placed into the same tube. 0.75 ml PE buffer was added to the Qiaquick column and centrifuged for 30-60 seconds. The Qiaquick column was placed in a clean 1.5 ml micro centrifuge tube and the DNA was eluted with 40 μl elution buffer.
The purified PCR products were cloned using QIAGEN PCR cloning plus kit as described by the manufacturer. Clones were selected and isolated plasmids with insert were sequenced with M13 Sequencing Primers using ABI biosystems automated sequencer (Macrogen Genomics, Korea).
Nucleotide database was searched with the sequences obtained using NCBI BLAST (Blastn) tool [13].
Sequences of both the control and treated Phaseolus and Vigna species were analyzed using pairwise sequence alignment LALIGN software from EMBNET server [14].
The total RNA isolated from P. vulgaris L. and V. unguiculata L. (cold stress treated and control) plants produced a distinct band and it was observed in 1.2% FAGE on UV illumination (Figure 1 and 2). Discrete band for P. vulgaris L. control dehydrin had the size of 911 bp, while treated dehydrin had 937 bp. In V. unguiculata L. control dehydrin (Dhn1) mRNA had 908 bp, while treated dehydrin (Dhn1) mRNA had 982 bp, in 1% agarose gel on UV illumination after loading the RT-PCR product amplified using Dhn primers.
The amplified products were purified and sequenced. The sequences were obtained from automated sequencer. The cold stress induced dehydrin gene of P. vulgaris L. and V. unguiculata L. and database sequence of control P. vulgaris L. and V. unguiculata L. plants were genetically analysed using Bioinformatics tools. Pair wise alignment indicated 98% similarity between the cold stresses induced dehydrin gene of P. vulgaris L. and V. unguiculata L. and database sequence of control P. vulgaris L. and V. unguiculata L. This genetic analysis concluded that there was no genetic variation between the LNT stress induced P. vulgaris L. and V. unguiculata L. plant when compared with database sequence of control P. vulgaris L. and V. unguiculata L.plants.
Lalign output for P. vulgaris L. control vs. P. vulgaris L. treated [ISREC-Server] Date: Fri May 4 4:24:26 Europe/Zurich 2007 LALIGN finds the best local alignments between two sequences version 2.0u66 September 1998 resetting to DNA matrix [14] Comparison of: (A) ./wwwtmp/lalign/.4415.1.seq Phaseo cont - 911 nt (B) ./wwwtmp/lalign/.4415.2.seq Phaseo test - 937 nt using matrix file: DNA, gap penalties: -14/-4 92.2% identity in 932 nt overlap; score: 3833 E (10,000): 3.4e-311.
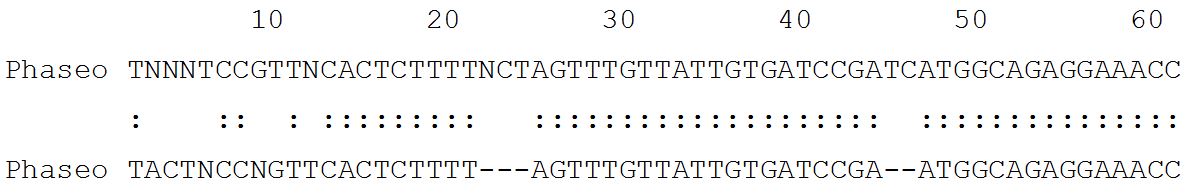
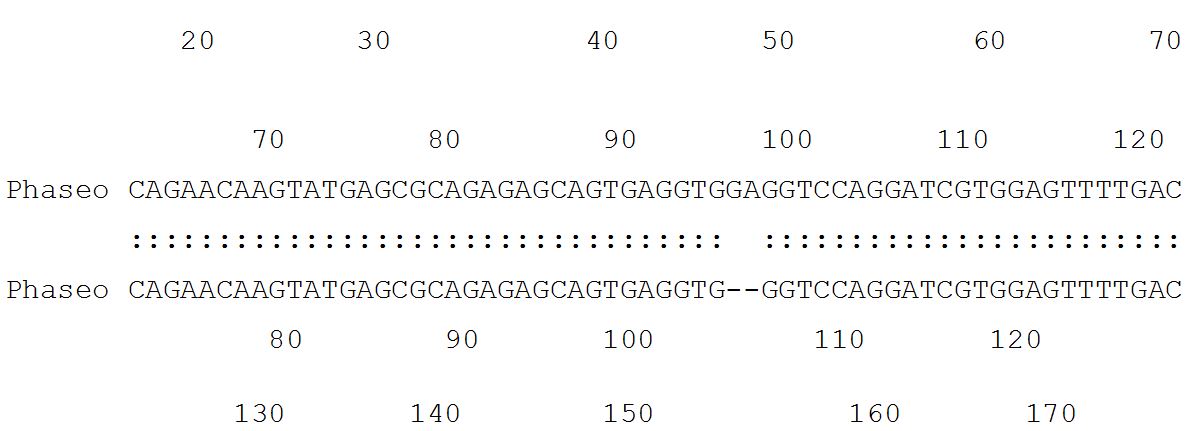
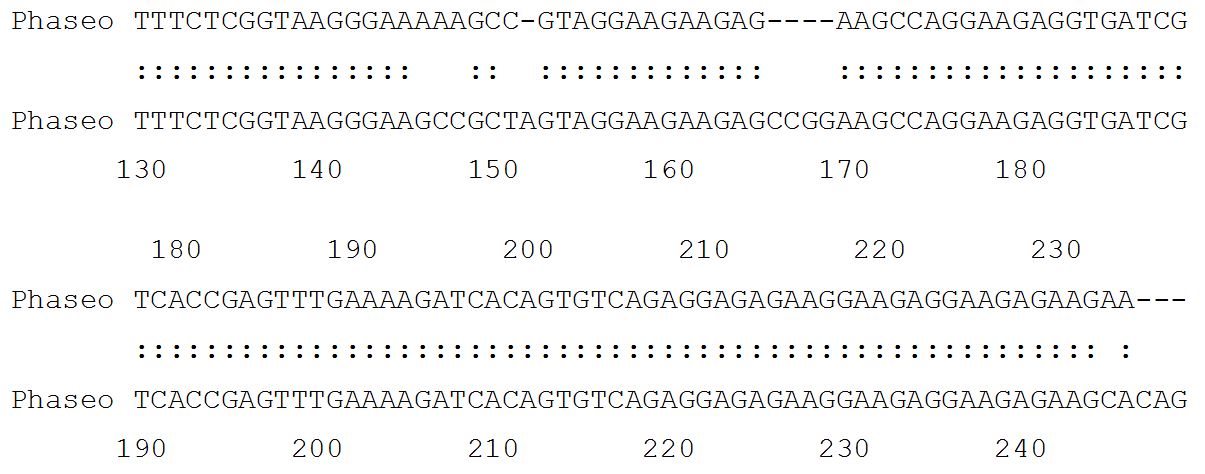
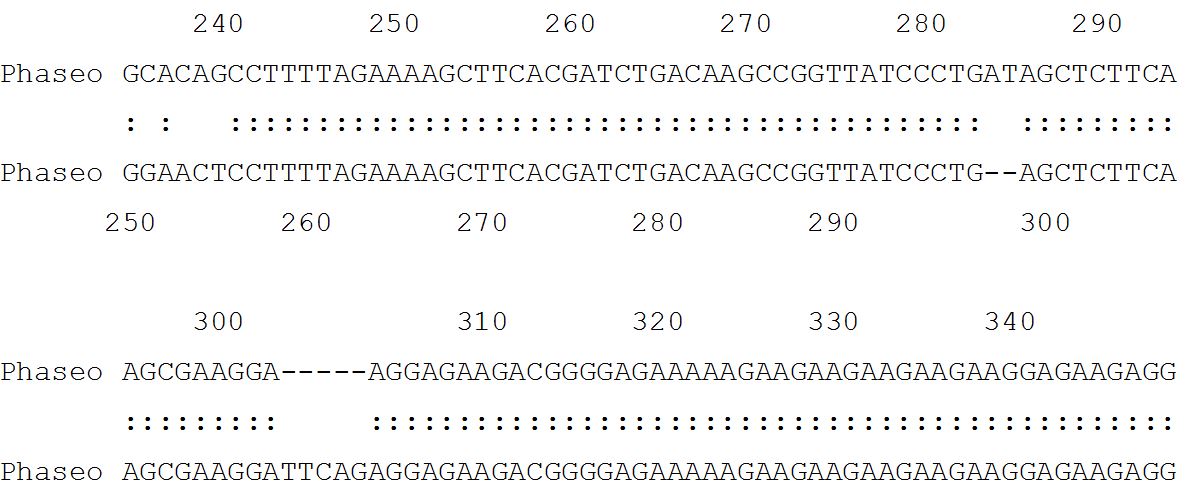
54.7% identity in 802 nt overlap; score: 383 E(10,000): 1.6e-23
55.4% identity in 578 nt overlap; score: 258 E(10,000): 4.3e-13
Lalign output for V. unguiculata L. control vs. V. unguiculata L. treated [ISREC-Server] Date: Fri May 4 4:14:27 Europe/Zurich 2007 LALIGN finds the best local alignments between two sequences version 2.0u66 September 1998 resetting to DNA matrix [14] 381 resetting to DNA matrix Comparison of: (A) ./wwwtmp/lalign/.4659.1.seq Vigna cont - 908 nt (B) ./wwwtmp/lalign/.4659.2.seq Vigna test - 982 nt using matrix file: DNA, gap penalties: -14/-4 96.0% identity in 916 nt overlap; score: 4137 E(10,000): 0
66.1% identity in 599 nt overlap; score: 868 E(10,000): 6.1e-64
61.3% identity in 802 nt overlap; score: 865 E(10,000): 1.1e-63
A growing number of genes have been shown to be induced during cold acclimatization. The cold stress induced dehydrin gene of P. vulgaris L. and V. unguiculata L. and database sequence of control P. vulgaris L. and V. unguiculata L. plants were genetically analysed using Bioinformatics tools. Pair wise alignment indicated 98% similarity between the cold stress induced dehydrin gene of P. vulgaris L. and V. unguiculata L. and database sequence of control P. vulgaris L. and V. unguiculata L. Dehydrin gene enhances tolerance to freezing stress in Arabidopsis and in Transgenic Arabidopsis plants over expressing multiple dehydrins were generated [7,15]. These findings showed that over expression of dehydrin results in increased freezing tolerance. This genetic analysis concluded that there was no genetic variation between the LNT stress induced P. vulgaris L. and V. unguiculata L. plant when compared with database sequence of control P. vulgaris L. and V. unguiculata L. plants.
This observation hypothesizes that there is no significant gene level variations between LNT stresses induced plants and control plants. It can also be hypothesized that variations may be occurring in its expression level to code for the protein responsible for making the plant tolerant to cold. Dehydrin gene produces the protein in response to LNT stress [16]. Moreover the present study also suggested no significant gene level variations between P. vulgaris L. and V. unguiculata L. plants even though significant biochemical variations were observed when these plants were exposed to LNT stress.
In this study, we have attempted to study the response of dehydrin gene in P. vulgaris L. and V. unguiculata L. to LNT stress. From this study, it can be inferred that dehydrin gene is induced to express the protein which confers an adaptation effect only in P. vulgaris L. but not in V. unguiculata L. LNT does not show any genetic variations when compared with a non LNT stress induced plant. Hence P. vulgaris L. is considered to be cold tolerant plant and V. unguiculata L. is a sensitive plant. P. vulgaris L. and V. unguiculata L. (LNT treated and control) can be analysed for other genes involved in cold stress. The Dhn gene can be cloned and expressed. The expressed protein characteristics can also be studied. The Dhn gene can be genetically transferred to plants using gene transformation methods to produce stress tolerant plants. This will be very useful for agriculturists to increase crop productivity which will be a great boon for a country like India with agriculture as prime occupation.