Isolation of Staphylococcus aureus from Ice-Cream Samples
Milk and dairy products including ice cream are good media for growth of Staphylococci, and dairy products are common sources of Staphylococcal intoxication. So the aim of this study is to detect the presence of Staphylococcus aureus in ice-cream samples which can be achieved by the following, 100 ice-cream samples were examined for presence of S.aureus on mannitol salt agar, the suspected colonies were identified by Gram staining and biochemically through catalase, oxidase and coagulase tests. Molecular identification of S.aureus by polymerase chain reaction (PCR) through detection of 16SrRNA gene and clfA gene specific for S.aureus. Antimicrobial susceptibility testing were carried out through disc diffusion method to detect the susceptibility of all isolates to various antibiotics and molecular detection of mecA gene. This study showed that 22 out of 100 ice-cream samples were appearing positive Staphylococci through conventional methods of isolation. 15 samples were positive S.aureus according PCR technique. PCR detection of mecA gene showed that 10 out of 15 (66.6%) isolates have mecA gene. Disc diffusion method showed most of Staphylococcus aureus isolates were multi antibiotic resistant (MAR) to oxacilin, penicillin, rifampin and nalidixic acid but they were sensitive to chloramphenicol, vancomycin and tetracycline.
Keywords: Ice-Cream; Staphylococci; Antibiotic; mecA and clfA
Ice cream is one of the dairy desserts, it is a popular frozen food consumed particularly in summer, as well as, throughout all the year. It continues to present a dominant interest for a large segment of population. Many studies from different countries revealed that ice-cream also acts as a vehicle of food-borne diseases [1-3].
The Staphylococci are ubiquitous in nature, with humans and animals as the primary reservoirs. They are present in the nasal passages and throat, in the hair, and on the skin of probably 50% or more of healthy individuals. These organisms are associated with sore throats and colds, and are found in abundance in postnasal drip following colds. Staphylococci can be isolated from animals, with the bovine being the most important because of the involvement of Staphylococci in mastitis. Although animals and humans are the major source, Staphylococci also can be found in the air, dust, water, and human and animal wastes [4].
Milk and dairy products are excellent growth media for a large number of microorganisms, including Staphylococci [5]. Bacterial contamination of milk usually occurs during the milking process and this depends on the sanitary condition of the environment and utensils used for milking and the milker’s hands, also it can gain access to milk by direct excretion from udders with clinical or subclinical staphylococcal mastitis [6,7].
Staphylococcus is one of the major bacterial pathogens which cause food poisoning [8]. Staphylococcal food poisoning (SFP) is a mild intoxication occurring after the ingestion of food containing Staphylococcal enterotoxins (SEs) [9]. There were five major classical SEs types, named; SEA, SEB, SEC, SED, and SEE. But now, new genes encoding enterotoxin such as SEG to SEU are identified. One or more of these genes are thought to be involved in Staphylococcal food poisoning [10].
Antimicrobial resistance is an important public health concern worldwide. The development of resistance both in human and animal bacterial pathogens has been associated with the extensive therapeutic use of antimicrobials or with their administration as growth promoters in animal production [11]. Staphylococci have been reported to frequently show multiple antimicrobial resistance patterns [12]. This may be due to the indiscriminate use of antibiotics has led to the development of multiple antibiotic resistances thereby rendering the antibiotic treatment ineffective [5]. The utilization of antibiotics in periods shorter than the recommended can also contribute to the antibiotic resistance.
Multiple antibiotic resistant Staphylococcus aureus (S.aureus) strains have been isolated from milk obtained from cattle, beef and human samples in many parts of the world [13, 14]. The prevalence of antibiotic resistance usually varies between isolates from the different sampled stations and even between isolates from different herds on the same farm [15]. There is many studies were evaluated the S.aureus in Ice cream, the difference between them and this manuscript was comparing the conventional and molecular methods in detection of S.aureus.
A total 100 samples of traditional ice cream were collected from different markets in Qena city with different flavor (17 with vanillia, 19 with chocolate, 13 with strawberry, 14 with mango, 14 with bananas and 23with mixed taste) within the shelf life period .The samples were transported to laboratory in sterile and cold containers (4 °C) and preserved at this temperature. The samples were processed immediately upon arrival using aseptic techniques [16].
Isolation of Staphylococci
One gram of each sample was diluted with 9 ml of 1% buffered peptone water and homogenized in a stomacher for about 10 minutes [17].The diluted samples were plated onto mannitol salt agar (MSA). The plates were incubated aerobically at 37 °C for 18-24h. Characteristic Staphylococci colonies were further purified by sub-culturing onto MSA plates and the plates were incubated aerobically at 37 °C for 18 h–24 h. These isolates were retained for further bacterial identification [18].
Smears from the purified colonies were stained with Gram’s stain and examined microscopically under oil immersion lens [19]. The typical colonies were showed gram-positive cocci occurring in bunched, grapelike irregular clusters were taken as presumptive Staphylococcus species.
Biochemical tests were performed to confirm S.aureus using Catalase test, Oxidase test and Coagulase test [20]. All S.aureus isolates were positive for catalase and coagulase and all of them were negative for oxidase test.
In this study PCR technique was carried to confirm the presence of S.aureus in 22 ice-cream samples which selected by detection of 16S rRNA gene specific for Staphylococci and clfA gene specific for S.aureus from culture samples Through the following steps [21].
It was carried out according to QIAamp DNA Mini Kit instructions (Catalogue no.51304).
PCR Amplification:For PCR amplification, using specific primers for each gene as shown in Table 1. A uniplex reaction mixture 25μl contained 1μl of Forward primers, 1μl of Reverse primers for each gene separately; 12.5μl Emerald Amp GT PCR master mix (2x premix), 6μl Template DNA and 4.5μl PCR grade water. The tubes were subjected to thermal cycling (Biometra) with programme described as shown in Table 2. Amplified products were separated by agarose gel electrophoresis (1.5%) agarose containing 0.5 mg ethidium bromide per ml (sigma) Visualized and photographed under UV illumination. The sizes of the amplication products were estimated by comparison with a 100 bp DNA ladder (Fermentas. cat. no. SM0243).
Antimicrobial susceptibility tests were performed by disc diffusion method according to the guidelines of Clinical and Laboratory Standard Institute [23]. Sensitivity pattern of the isolates were determined against Oxacillin (1mcg), Vancomycin (30mcg), Cefotaxime (30mcg), Chloramphenicol (30mcg), Nalidixic acid (30mcg), Rifampin (5mcg), Penicillin G (10mcg) and Tetracycline (30mcg). Antimicrobial testing results were recorded as sensitive, intermediate sensitive and resistant according to zone diameter interpretative standards provided by [23].
A uniplex reaction mixture 25μl contained 1μl of Forward primers 1μl of Reverse primers for each gene separately; 12.5μl Emerald Amp GT PCR master mix (2x premix), 6μl Template DNA and 4.5μl PCR grade water. The amplification was carried out with the following conditions: 94 °C for 5 min as Primary denaturation, 94 °C for 30 sec denaturation, annealing at 50 °C for 30 sec, extension at 72 °C for 30 sec and final extension at 72 °C for 7 min for 35 cycles.
According to conventional method of identification through culture on mannitol salt agar, microscopic and biochemical identification, they were 22 samples positive for S.aureus. They have yellow colonies on mannitol salt agar; microscopically appear gram positive cocci, arranged in clusters, non-spore forming bacteria, positive catalase test and negative oxidase. Coagulase test showed that 4 isolates were strong coagulase and 8 isolates were suspected coagulase (weak coagulase).
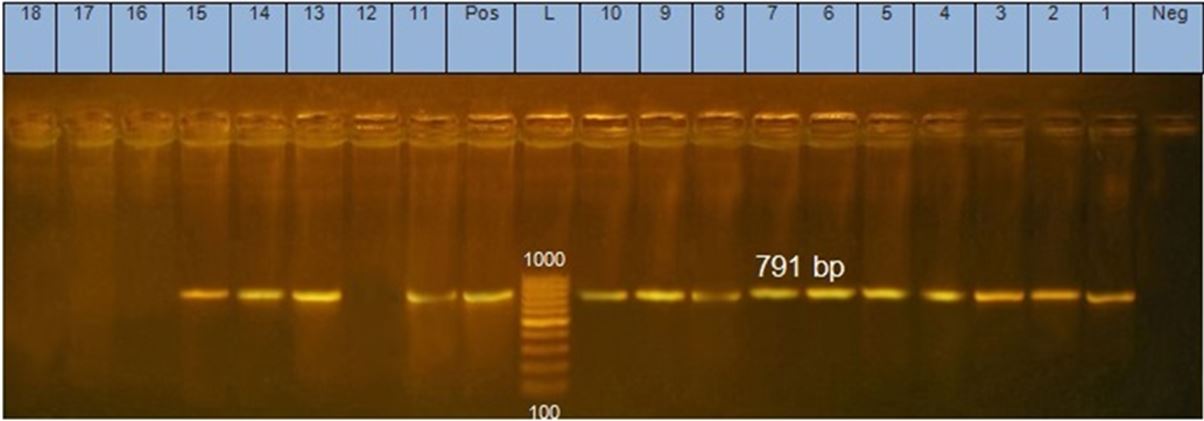
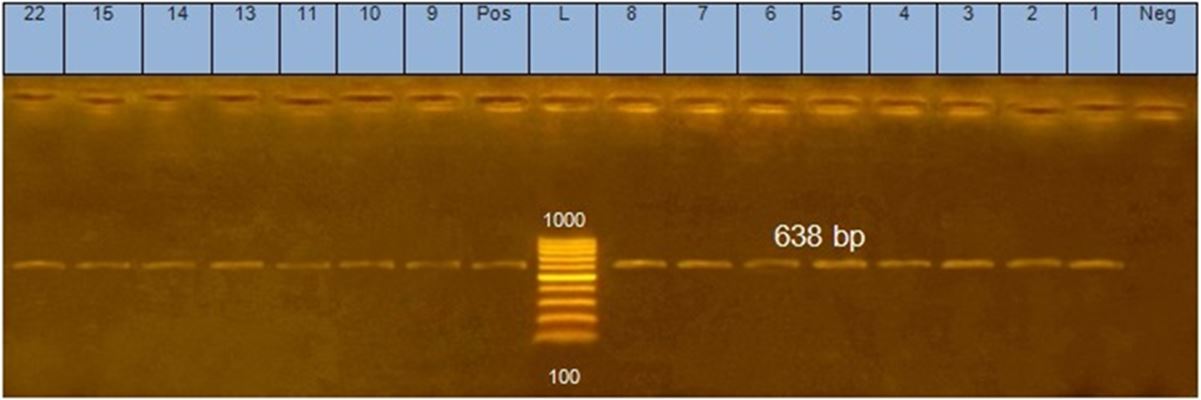
All 22 isolates were examined by PCR using 16S rRNA gene specific for genus Staphylococcus and clfA gene specific for S.aureus. 15 out of 22 isolates were positive Staphylococci as shown in Figure 1, and all of them were S.aureus according clfA gene result as show in Figure 2.
All the 15 isolates were examined for antibiotic sensitivity to different antibiotics through disc diffusion method. Antimicrobial susceptibility testing through disc diffusion method shows the following (Table 3):
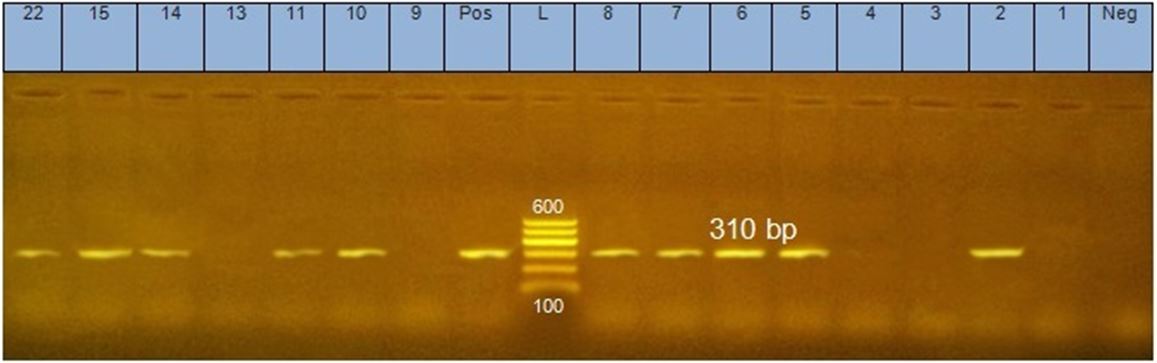
Among the 15 S.aureus strain, there were 10 strains have mecA gene, as show in Figure 3.
Ice cream is one of the most abundant and popular dairy products that are consumed in warm seasons by vulnerable groups, especially children, therefore; its microbial contamination is very important. This study has shown contamination of ice-cream with S.aureus.
The present study aimed to detect S.aureus and detection of antibiotic resistance profile and methicillin resistance gene of S.aureus isolated from ice cream.
A total 22 (22%) S.aureus isolates were detected from 100 samples of ice cream, all the isolates were gram positive cocci arranged in clusters, catalase positive and oxidase negative which agree with who isolated S.aureus from 11/50 (22%) ice-cream samples. And nearly similar finding to that assumed by who isolated S.aureus from 26%, 22.9% and 20%, respectively [25-28].
On the other hand, higher incidence was reported by who isolated S.aureus from 84.72%, 50%, 76%, 56.67% and 55% respectively, but lower incidence was reported by isolated S.aureus from 2.7 %, 4.3%, 0.5%, 12.2% and 4.4%, respectively. Moreover, could not detect S.aureus in any one of the examined ice cream samples. The difference in the incidence may be due to bad hygienic measurement during manufacture of ice cream [29-42].
In this study the 22 S.aureus isolates were conducted for PCR and we found 15 out 22 (68.2%) showing positive using 16SrRNA gene and all of them were S.aureus using clfA gene specific for S.aureus which in agreement with because the PCR is highly sensitive while conventional method is less sensitive as there is some microorganism give positive reaction by culture and biochemical tests but give negative by PCR [21].
Antimicrobial resistance is an important public health concern worldwide. It has been believed that all bacterial infections treated with effective antimicrobial agents. However, the emergence of resistance to multiple antibiotics among S.aureus has created breaking news for health practitioners and researchers [43]. It has been reported shortly after introduction of penicillin 1940s, resistance developed in S.aureus followed by resistance to methicillin and more recently to glycopeptides as vancomycin [44].
In this study the isolates were evaluated for antimicrobial resistance through disk diffusion method, resistance to penicillin G was high (93.3%), high resistance to β-l actam antibiotic was not surprising as it is commonly used for treatment of infections in humans and animals [45].
The present study S.aureus was resistant to oxacillin (80%), this result nearly similar to and who found resistance of S.aureus to oxacillin 76.2% and 86.7% respectively. While, this result disagreed with that of and who detected resistance rate against oxacillin of 6.2% and 28% respectively [46-49].
This study show that one S.aureus isolate sensitive to oxacillin and contain mecA gene which nearly similar to who obtained results demonstrated low correlation (p>0.05) between phenotypic resistance to oxacillin and the presence of mecA gene in Staphylococci and found the similar results in phenotypically oxacillin resistant isolates of S. aureus. Those strains did not carry neither mecA nor mecC genes [50,51].
Furthermore, there were four isolates show oxacillin resistance phenotypically and could not have mecA gene which agree with who found only 5 isolates have mecA gene out of 22 isolates showing oxacillin resistance phenotypically by disc diffusion method. Oxacillin has been proposed as a proxy antibiotic for testing susceptibility not only to methicillin and to all β-lactams, which could explain why all oxacillin-resistant isolates were not carrying the mecA gene [52,53].
The present study shows that S.aureus isolates were resistant to vancomycin 6.6%.This nearly agree with who found Staphylococcus isolates were resistant to vancomycin with 3.33% and disagree with who reported that S.aureus were 100% resistant to vancomycin [54,55].
The present study show that S. aureus isolates were sensitive to chloramphenicol 93.3% which nearly agree with who found 85.96% of S.aureus isolates sensitive to chloramphenicol and who reported that all S.aureus isolates were susceptible to chloramphenicol. These results were not in agreement with and who found resistance of S.aureus isolates to chloramphenicol 33.3% and 25% respectively. Also, it disagree with who found that S.aureus were resistant to chloramphenicol in 80% [46, 55-57].
The resistance to tetracyclines occurs through the presence of tet genes in the bacterial DNA. The characterized tet genes encode three mechanisms of resistance: efflux pump, ribosomal protection or enzymatic inactivation 28 [59].
In this study, 73.3% of the isolates were susceptible to tetracycline which nearly agree with who found about 75.44% of the isolates were sensitive to tetracycline. And disagree with and who reported that S.aureus were resistant to tetracycline in 74.1% and 85% respectively. Also it disagrees with who observed that all isolates of S.aureus (100%) were resistant to tetracycline [4, 46,56,58].
About 53.3% of the isolates were resistant to rifampin which were disagree with who reported that 68.4% of S.aureus isolates were sensitive to rifampin [56].
The sensitivity of S.aureus isolates to cefotaxime is (53.3%) which were not in agreement with who reported that, all S.aureus isolates were susceptible to cefotaxime [57].
In this study, the isolates were resistant to nalidixic acid in 60% which were not in agreement with who found that all S.aureus isolates 100% were resistant to nalidixic acid. The difference in resistance of Staphylococcus aureus isolates to different antibiotics may be due to genetic variation and phenotypic variation.
It was observed that 66.6% of S.aureus isolates were positive for presence of mecA gene and this in agreement with who found 66.7% of S.aureus isolates were positive for presence of mecA gene and who isolate MRSA in 50% and disagree with and who detect MRSA in 18.18% and 29.6% respectively [46, 61-63].
Ice cream is one of most popular and favorite food products all over the world. It is an ideal media for microbial growth due to high nutritive value and long storage duration. Once the ice becomes cream contaminated, freezing temperature could not make the product safer later.
This study revealed that, some ice cream sold in Qena city, Egypt was contaminated with Staph. aureus which may cause food poisoning. It indicates the lack of hygienic conditions during preparation and preservation of ice cream. Antimicrobial resistance is an important public health concern worldwide. In the present study, Staph.aureus isolates exhibit resistance towards the different antibiotics tested.
Thorough food inspection and frequent bacteriological surveillance by food control agencies is highly recommended to control the incidence of Staph.aureus in dairy products to safeguard the consumers from risks of food poisoning.