Preparation and Characterization of Biodegradable Glimepiride Loaded PLA Nanoparticles by o/w Solvent Evaporation Method Using High Pressure Homogenizer: A Factorial Design Approach
Glimepiride is used for the treatment of type 2 diabetes mellitus, one of the third generation sulfonylurea drugs having poor aqueous solubility, slow dissolution rate and low elimination half life. The Aim of the present study was to prepare Glimepiride loaded poly-D, L-lactide (PLA) Nanoparticles by o/w solvent evaporation method using high speed homogenizer. Prepared Nanoparticles were characterized for the drug content, encapsulation efficiency. Physical state of the drug, polymer and Nanoparticles were determined by Particle size distribution, differential scanning calorimetry (DSC), X- ray diffraction (X-RD), Fourier transform infrared spectroscopy (FTIR). Particle morphology was observed by FE-SEM. The results of this study showed the potential of the O/W emulsion for drug delivery application. Drug content and Encapsulation efficiency were found to be 40.27 and 80.55% respectively and particle size of the Nanoparticles was 442 nm (dnm). DSC and XRD study showed partial interaction between the drug and polymer.
Drug release of optimized batch showed 73.72% to 78.12% drug released up to 12 hrs and optimized formulation followed first order kinetics. From this study we can conclude that the Glimepiride loaded PLA nanoparticles were successfully prepared and these nanoparticles seem to be promising for sustained drug release application leading to improved patient compliance.
According to the biopharmaceutical classification system, approximately 40% of drugs in the industry pipeline belongs to the BCS class II having low aqueous solubility and high permeability and class IV having low solubility and low permeability. Drugs belong to these classes have low bioavailability [1-3]. Therefore, these drugs possess formulations and delivery problems [4]. For the treatment of diabetes, type 2, the use of oral anti-diabetic drugs increases rapidly [5]. Glimepiride is used for the treatment of type 2 diabetes, one of the third generation sulfonylurea drug having a poor aqueous solubility, slow dissolution rate and low elimination half-life (2-3 hrs) [6]. Glimepiride have a number of advantages over other members of sulfonylurea, currently in the market such as lower dosages, fast onset of action and lower C-peptide level of insulin, this is because of less secretion of insulin and more pronounced extra pancreatic effects [7]. Glimepiride acts by binding to the specific site on pancreatic β-cells and block the ATP-Dependent potassium channels to stimulate the insulin release. Due to the short elimination half-life frequent dosing is required, which leads to the adverse effects such as headache and gastrointestinal disorders [8].
Due to its short elimination half-life repeated doses are required which may cause different side effects to avoid the repeated dosing and enhance the bioavailability of glimepiride sustained release nanoparticles were developed. Nanotechnology is promising application in drug delivery system that accounts for the main part of nanomedicine [9]. There are number of biocompatible polymers are available in the market such as ethyl cellulose, PLA, poly (lactic-co-glycolic acid) (PLGA), poly-ε-caprolactone (PCL) and poly glycolic acid (PGA) for the formulation of sustained as well as controlled release nanoparticles [10]. Of these, PLA is more suitable polymer for sustaining the drug release and is used for the encapsulation of many therapeutic agents due to their biocompatibility, biodegrability, low toxicity, high mechanical strength and slow drug release [11].
Factorial analysis (FA) has been widely adopted for optimization of various parameters in drug formulation and development. This is a collection of statistical and mathematical techniques useful for developing, improving, and optimizing processes in which a response of interest is influenced by multiple variables and the objective is to optimize this response. Optimization of drug encapsulation efficiency by utilizing FA may allow more comprehensive analysis of the interactions between the experimental variables than a single-factor or two factor experimental designs [12,13]. Consequently, this could lead to a better understanding. Apart from that, it also reduces the number of experimental runs required to generate statistically-validated results [14,15].
In recent years, several approaches have been investigated for the development of nano range drug delivery system. Method of preparation of nanoparticle can be classified into the top-down and bottom up processes [3]. Top-down process involves size reduction of the larger particle into the micrometre or nano range. In this process size reduction of the particle can be achieved by milling, high pressure homogeniser or micro fluidization [16] where as in bottom up process, the drug nanoparticles are formed from molecules in a solution or in an emulsion. Emulsion evaporation, solvent displacement, solvent diffusion and rapid freezing have been widely used to prepare drug nanoparticles. Nanoparticle produced by top down process are generally crystalline in nature and require high energy and high pressure for the development of nano size particles, there is a chance of contamination if a milling medium used. Whereas bottom up process involves dissolution followed by precipitation or drying [17-19].
In the present study Glimepiride loaded PLA Nanoparticles were prepared by an o/w solvent evaporation method using a high pressure homogenizer to overcome the dosing frequency and side effects.
Glimepiride was obtained as a gift sample from Wockhardt Research Centre, Aurangabad, Maharashtra, India. Poly-D, L-lactide (PLA) was purchased from Merck specialties Pvt. Ltd. Dichloromethane (DCM) and Acetone were purchased from Merck specialties Pvt. Ltd. and RFCL limited, India respectively. Poly vinyl alcohol was purchased from S.D fine-chem. Ltd, India. All other chemicals and reagents were of analytical grade and used as provided.
Factorial design: A number of preliminary experimentations were conducted to determine the formulation parameters and conditions at which the process to yielded. To optimize the formulation a 32 full factorial design was applied for the preparation of inclusion complex using Design-Expert® Software (Version-8.0.7.1, Stat-Ease Inc., Minneapolis,) which allows evaluation by nine experiments in order to limit the number of experiments. Response surface methodology (RSM) was used for the analysis of results. The amount of PLA used as release retarding polymer (X1,mg) and PVA as a surfactant (X2, %w/v) with respect to drug was selected as independent variables. Such statistical models were used to evaluate the effect of independent variables on the dependent variables like encapsulation efficiency (Y1, %), practical drug loading (Y2, %).The actual and coded values of independent variables are shown in Table 1 along with their low (−1), medium (0) and high levels (+1). The significance of the model was determined by the comparisons of statistical parameters, and the best model (suggested) was decided based on reasonable agreement between adjusted R2 and predicted R2 ; higher values of adjusted R2 and predicted R2; model p value (should be less than 0.05). Two-dimensional (2D) contour plots and three-dimensional (3D) response plots resulting from the equations were constructed using Design-Expert® software [19,20].
Preparation of nanoparticles: Nanoparticles containing anti-diabetic drug as a core material were prepared by O/W solvent evaporation method [21,22]. Accurately weighed quantity of drug (50mg) was dissolved in DCM & acetone (5 ml each) and polymer in DCM (10 ml) separately and added into the aqueous phase (100 mL distilled water) containing surfactant 0.45% PVA using a high speed homogenizer (OMNI International TH) at 20000 rpm for 20 min and 35000 rpm for 3 min at 4 oC, by adding organic phase into aqueous phase. Then emulsion was passed through the high pressure homogenizer at a pressure of 500 bar for one cycle. The emulsion was kept on lab stirrer (Remi, Mumbai) at 1000 rpm for 3 hrs at room temperature for the evaporation of organic solvent, after that the nanoparticles were collected by centrifugation for 5 minutes at 10000 rpm (Remi, Mumbai). PVA is a water soluble surfactant, during centrifugation it was removed along with decant. Trace amount of PVA present in nanoparticles were removed by washing with distilled water. After washing nanoparticles were lyophilized for 48 hrs. To minimize the potential risk in patients treated with sustained release glimepiride the lowest effective dose should be used for the shortest possible duration [19].
The amount of Glimepiride encapsulated into nanoparticle was determined by UV spectrophotometer (HITACHI U-2900, Tokyo, Japan). An accurately weighed 10 mg of nanoparticles were stirred with dichloromethane (5 ml) to dissolve the polymeric coat and extracted in phosphate buffer solution (pH 6.8). Stirring continued for 30 min at 30-40 °C to facilitate an evaporation of organic solvent. The drug content was determined in the filtrate after appropriate dilution with phosphate buffer solution at 226 nm using UV-spectrophotometer. The drug content and encapsulation efficiency of Glimepiride were calculated by using the following equations:
% Drug Loading = [drug determined/ actual drug added] × 100 - (I)
% Encapsulation efficiency = [actual quantity/theoretical quantity] × 100 - (II)
Thermal behavior of the sample was determined by Differential Scanning Calorimetry (DSC-60, Shimadzu & 821, Mettler Toledo). Accurately weighed samples (5-10 mg) were sealed in an aluminium pan and scanned at a temperature range of 30 °C to 400 °C at the rate of 10 °C/min under dry nitrogen atmosphere purge of 50 mL/min.
X-ray diffraction analysis of sample was carried out to characterize the physical form i.e. amorphous or crystalline nature of glimepiride in sample of optimized batch in an X-ray diffractometer (D8 Advance, Bruker) with Cu Kα radiation (λ=1.54060 A°). The scanning rate 100/min and diffraction angle 2Ø was 10-80°.
The morphology of the samples was carried out using scanning electron microscopy (FE-SEM, Type-II, Model-S-4800, Hitachi, Japan).The surface morphology was analyzed at a working distance of 8.7-8.8 mm and 1.0 KV accelerating voltage.
The particle size distribution and zeta potential was determined in water as a dispersion medium by laser diffraction size analyzer, Malvern Zetasizer (Model: ZS 200).
Drug polymer interactions were studied by using an FTIR spectrophotometer (Shimadzu, FTIR-8400). FTIR analysis of the pure drug, polymer and nanoparticles of glimepiride was carried out by KBr pellet method. The sample was mixed with KBr and compressed into a disc in a manual press. The spectrum was scanned from 4000 to 400 cm-1.
Drug release studies of the nanoparticles were performed, in a Tablet Dissolution Test Apparatus, Type-II (Paddle method, Electrolab, TDT 06) at 37±0.5 oC and at a paddle speed of 75 rpm. The release studies were carried out in a 900 mL dissolution medium of pH 1.2 for the first 2hrs, and continued in phosphate buffer pH 6.8 up to 12 hrs. Change in pH was made by the addition of 0.2M tribasic sodium phosphate. All Dissolution media was contained 0.2% Sodium lauryl sulfate. 5 mL sample was withdrawn from the dissolution apparatus at different time intervals and filtered through membrane filter. The drug content was determined at 226 nm by Double beam ultraviolet spectrophotometer. The withdrawn sample was replenished with 5 mL of fresh media.
To study the drug release mechanism from sustained release nanoparticles the release data were fitted to zero-order kinetics, first-order kinetics [19], Higuchi model and Korsmeyer-Peppas kinetics models [23]. The kinetic model with the highest value of the coefficient of correlation (r2) was considered to be a best fit model for describing the release of glimepiride from the nanoparticles [24].
Using 32 factorial design, a total of nine runs was carried out for the preparation of inclusion complex and investigate the effects of two independent variables on the dependent variables (responses) using factorial design. The quadratic mathematical model (suggested) generated by 32 factorial design was used to evaluate the responses.
Y = β0 + β1 X1 + β2 X2 + β3 X1 X2 + β4 X12 + β5 X22 (3)
Where, β0 is the intercept; β1 to β5 are the estimated coefficient obtained from the observed experimental values of Y; X1 and X2 are the coded levels of the factor. The coefficient corresponding interaction (X1X2) and the quadratic effects (X12 and X22) were determined from the results of the experiments. Results of all the nine experiments carried out are summarized in Table 2. A study showed that the formulation parameters had an influence on the encapsulation efficiency and solubility of FMT in complex. The polynomial model describing the correlation between the formulation variables and the response can be represented by the following equation.
Y1 = 73.22– 0.083967X1 +39.3X2 – 0.8575X1X2+ 3.2066X12 +1.0266X22 (4)
Y2 = 55.87– 0.43397X1 – 8.9166X2 – 0.171X1 X2 + 0.0012X12 + 102.66X22 (5)
| Experimental run order | Variables | Theo. DL(%) | |||
|---|---|---|---|---|---|
| Independent variables | Independent variables | ||||
| X1 | X2 | Y1 | Y2 | ||
| 1 | 50 | 0.15% | 65.22% | 32.50% | 50% |
| 2 | 50 | 0.30% | 70.37% | 35.38% | 50% |
| 3 | 50 | 0.45% | 80.55% | 40.27% | 50% |
| 4 | 100 | 0.15% | 61.72% | 20.505 | 33.30% |
| 5 | 100 | 0.30% | 63.95% | 21.20% | 33.30% |
| 6 | 100 | 0.45% | 66.40% | 22.27% | 33.30% |
| 7 | 150 | 0.15% | 52.96% | 13.24% | 25% |
| 8 | 150 | 0.30% | 56.28% | 14.07% | 25% |
| 9 | 150 | 0.45% | 61.11% | 15.37% | 25.00% |
Table 2: Drug Loading (DL) and Encapsulation Efficiency (E.E.) of Glimepiride Loaded PLA Nanoparticles
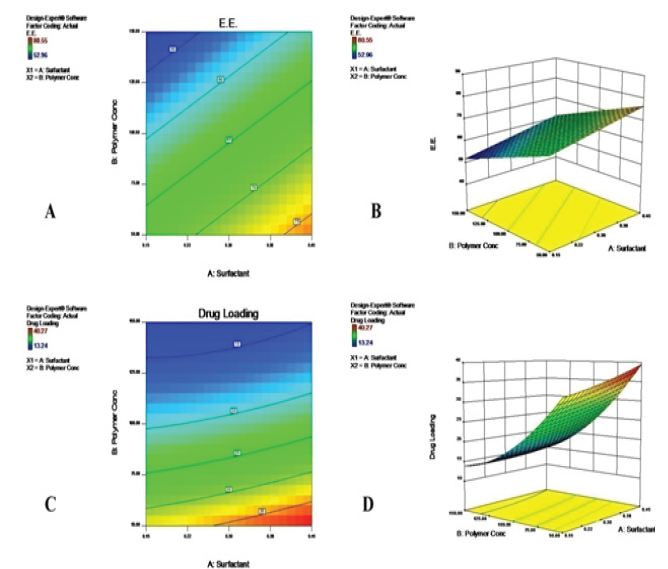
The equation represents the quantitative effects of factor (X1 and X2) upon the responses (Y1 and Y2). The sign of the coefficient shows how the factor influences the response. If the coefficient is positive, the response is increased (synergistic effect) as the factor moves from low level (−1) to high level (+1); the contrary is obtained (inverse relationship/antagonist effect) if the coefficient is negative. Linear, cross-product contribution (2FI), quadratic and cubic models were generated for the responses by the software. The quadratic model showed a best fit for the responses. Table 3 showed the model summary statistics of responses. Data in Table 4showed the coefficient estimate and p values of each factor for the measured responses. Significant values indicated in bold faces. Significant factors affecting the response Y1 were X1 (amount of PLAas a polymer p value 0.0009), X2 (amount of surfactant, p value 0.0058), and quadratic term X22 (p value, 0.050). Significant factors affecting the response Y2 were X1 (p value, <0.0001), X2 (p value, 0.0066), and quadratic term X2>2 (p value, 0.0097). To validate the model, all the points were selected and observed their experimental and predicted value for the responses. Therefore, it can be concluded that the model is best suitable because of the difference between experimental and predicted value is very low.
Drug loading and encapsulation efficiencyof different experimental runs of Glimepiride nanoparticles are given in Table 2. Glimepiride loaded PLA nanoparticles showed practical drug loading 40.27% and 80.55% encapsulation efficiency respectively which is shown in Figure 1. From the study it was observed that the increase in PVA concentration increases the %E.E and %DL but decreases with increase in polymer concentration. This may due to the decrease in interfacial tension with increase in PVA concentration and also increases the surface area of PLA nanoparticles, which leads to the more diffusion of the drug from the nanoparticles and it may decreases the %E.E., but in our findings it increased due to the free drug available on the surface of nanoparticles in place of entrapment on the nanoparticles. While %E.E. decreases with an increase in the amount of polymer concentration, this may be due to the more time taken for the precipitation of polymer which was in higher amount [25]. Among the nine formulations, Run R3 and R9 are the optimized formulations in terms of %E.E. and % DL.
| Response | Model | Standard deviation | R2 | Adjusted R2 | Predicted R2 | Significance |
|---|---|---|---|---|---|---|
| Y1 | Linear | 1.86 | 0.9613 | 0.8970 | 0.5296 | |
| 2FI | 1.73 | 0.9316 | 0.8906 | 0.7209 | - | |
| Quadratic | 1.68 | 0.9043 | 0.8725 | 0.7602 | Suggested | |
| Cubic | 0.11 | 0.9999 | 0.9995 | 0.9896 | - | |
| Y2 | Linear | 2.13 | 0.9635 | 0.9514 | 0.9139 | - |
| 2FI | 2.21 | 0.9675 | 0.948 | 0.8333 | - | |
| Quadratic | 0.76 | 0.9976 | 0.9937 | 0.9727 | Suggested | |
| Cubic | 0.47 | 0.9997 | 0.9976 | 0.9460 | - |
| Response | Y1 | Y2 | ||
|---|---|---|---|---|
| Coefficient of estimate | p value | Coefficient of estimate | p value | |
| X1 | -4.98 | 0.0009 | - 10.88 | <0.0001 |
| X2 | 3.93 | 0.0058 | 1.5 | 0.0066 |
| X22 | 3.93 | 0.0500 | 1.5 | 0.0097 |
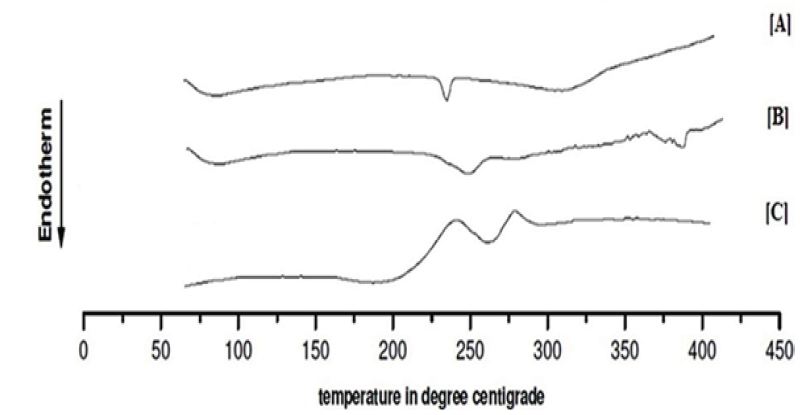
DSC thermo gram of pure drug, polymer and an optimized batch of nanoparticle are shown in Figure 2. Thermogram of pure drug showed a sharp endothermic peak at 234.13 0C which corresponds to melting point of the drug, which is same as reported in literature. A result of DSC analysis confirms the identity and purity of the drug. Glass transition temperature of the pure PLA polymer ranges from 150 to 160 oC. Pure PLA polymer showed Tg at 141.63 0C and Glimepiride loaded PLA nanoparticle show an endothermic peak at 262.48 oC during the DSC analysis. There was an increase in endothermic peak in nanoparticle as compared to pure drug, suggesting that the interaction between the drug and polymer occurs which is due to changes in physical form of crystalline to amorphous state [26].
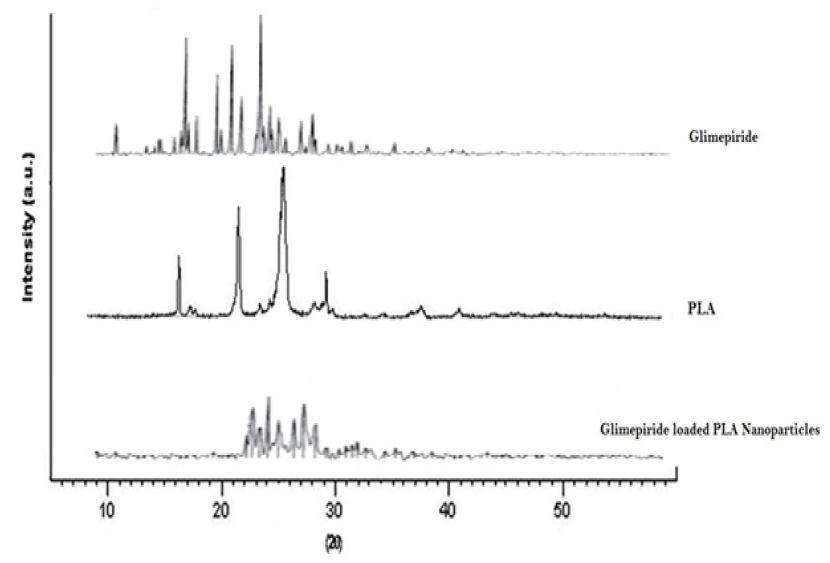
X-RD patterns of pure drug, polymer and nanoparticles were shown in Figure 3. The diffraction spectra of pure drug showed sharp and intense peaks of crystallinity. On the other hand X-RD pattern of nanoparticles prepared by the o/w solvent evaporation method showed reduction in number and intensity of the peaks in comparison to the pure drug indicates that the decrease in crystallinity or slightly change in amorphous nature of the drug [27].
The field emission scanning electron micrographs (FESEM) of the nanoparticle prepared by the o/w solvent evaporation method and were found to be spherical in shape Figure 4.
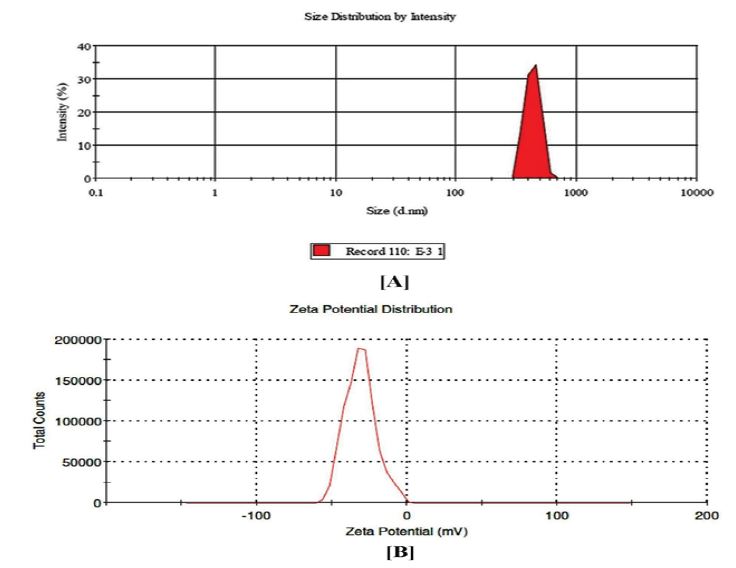
Particle size distribution and zeta potential of Glimepiride loaded PLA Nanoparticles formulation was found to be 442.2(d. nm.) and Zeta potential -31.2mV. Zeta potential value was found to be in the range of ±30 to ±40 in optimised formulation (R3 and R9) signifying an increase in stability of glimepiride loaded PLA nanoparticles. Result of particle size and Zeta potential distribution was shown in Figure 5a and 5b.
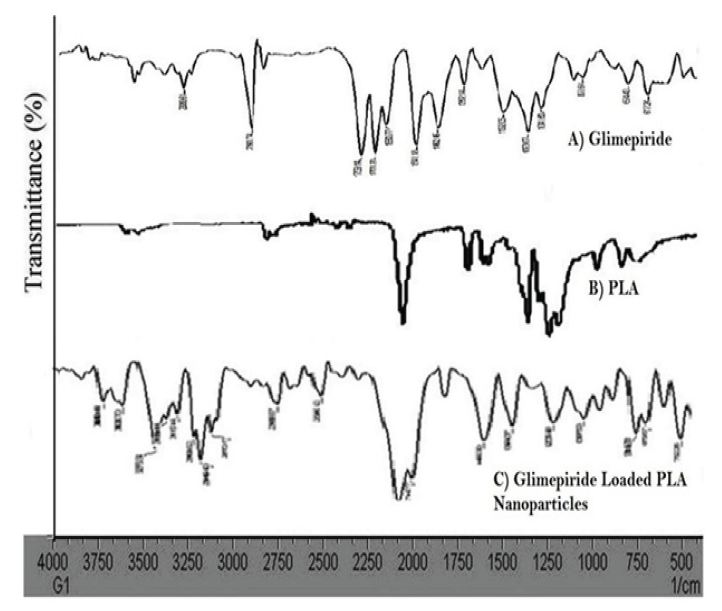
FTIR spectra of pure drug, polymer, and Glimepiride loaded PLA nanoparticles of optimized formulation are shown in Figure 6. FTIR spectra of pure drug showed characteristic peaks at 3369 cm-1 and 3288 cm-1 corresponding to N-H stretching, 1707 cm-1 and 1674 cm-1 due to carbonyl stretching, 1345 cm-1 showing C-N stretching, 1153cm-1 showing S=O stretching vibration. FTIR spectrum of different batches of glimepiride shows decrease in the intensity of N-H and S=O stretching vibration of glimepiride.
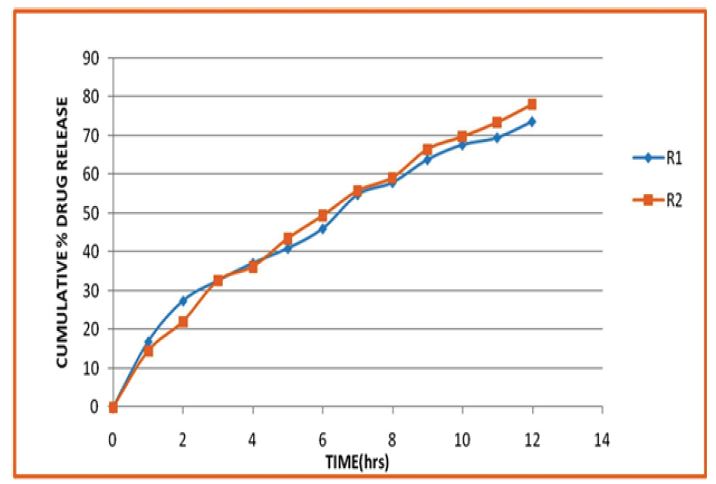
In vitro drug release studies of glimepiride loaded PLA nanoparticles were performed to determine the percentage of drug released from the nanoparticles in gastric fluid and intestinal fluid. The in vitro drug release study of optimized formulation (R3 and R9) with same concentration surfactants was performed for the first two hours at pH 1.2 and at phosphate buffer pH 6.8 for next 12 hrs. The release patterns of glimepiride from nanoparticles are shown in Figure 7. The drug released from the nanoparticles was found to be in the range of 14.46 to 16.89% in first 2 hrs at pH 1.2 and 73.72 to 78.12% up to 12 hrs at pH 6.8. From the drug release behavior it was observed that if the polymer concentration increase drug release decreases due to the high viscosity of PLA which on contact with the dissolution medium, surface of nanoparticles become wet and forms viscous gel layers. As the concentration of PLA increases viscosity of the gel layers increases while the diffusion coefficient of drug decreases [28,29].
To study the release behavior of glimepiride loaded PLA nanoparticles the release data were fitted to the various kinetic models such as a zero - order, first-order, Higuchi and Korsmeyer-Peppas kinetics models. In this present study in-vitro drug release profile of the optimized batch (R2) followed first-order kinetics and plot shows highest regression coefficient (r2 0.9915).The slope (n) value obtained from Korsmeyer-Peppus model was found to be less than 0.89 indicates drug releases by diffusion mechanism [30].
Glimepiride loaded PLA nanoparticles were successfully prepared by an o/w solvent evaporation method using a high pressure homogenizer with encapsulation efficiency of 80.55% and drug loading 40.27%. DSC and FT-IR study showed the partial interaction with drug and polymer due to the hydrogen bonding between N-H group and carbonyl group. A slow drug released from the glimepiride loaded PLA nanoparticles was observed and particle size 442.2 nm (denim) can make the suitable candidate for the further development of nano medicines.
The authors are thankful to Defence Research and Development Organization (DRDO)Govt. of India, New Delhi, for providing financial support, in terms of Extramural ResearchProject [ERIP/ER/0903820/M/01/1379].The authors also like to thank Wockhardt Research Centre, Chikalthana, Aurangabad, Maharashtra, India for providing the gift sample of Glimepiride and University Institute of Chemical Technology, North Maharashtra University, Jalgaon 425001, Maharashtra, India for providing all facilities to carry out this research work.
- Visser MR, Baert L, Klooster GV, Schueller L, Geldof M, et al. (2010) Inulin solid dispersion technology to improve the absorption of the BCS Class IV drug TMC240. Eur J Pharm Biopharm 74: 233–8.
- Keck CM, Muller RH (2006) Drug nanocrystals of poorly soluble drugs produced by high pressure homogenisation. Eur J Pharm Biopharm 62: 3–16.
- Chan HK, Kwok PCL (2011) Production methods for nanodrug particles using the bottom-up approach. Adv Drug Del Rev 63: 406–16.
- Chen H, Khemtong C, Yang X, Chang X, Gao J (2011) Nanonization strategies for poorlywater-soluble drugs. Drug Discovery Today 16: 354-60.
- Revena S, Grdadolnik J, Kristl J, Zagar E (2010) Hyperbranchedpoly (esteramides) as solubility enhancers for poorly water-soluble drug glimepiride. Int J Pharm 396: 119–126.
- Davis SN (2004) The role of glimepiride in the effective management of Type 2 diabetes. J Diabetes Complicat 18: 367–76.
- AmmarHO, SalamaHA, Ghorab M, Mahmoud AA (2006) Formulation and biological evaluation ofglimepiride–cyclodextrin–polymer systems. Int J Pharm 309: 129–38.
- Mriller G, Hartz D, Printer J, Okonomopulos R, Kramer W (1994) Differential interaction of glimepiride and glibenclamide with the/3-cell sulfonylurea receptor. I. Binding characteristics. Biochim Biophys Acta 1191: 267-77.
- Donga Y, Feng SS (2007) Poly (d,l-lactide-co-glycolide) (PLGA) nanoparticles prepared by high pressure homogenization for paclitaxel chemotherapy. Int J Pharm 342: 208–14.
- Lee LY, Wang CH, Smith KA (2008) Supercritical antisolvent production of biodegradable micro-and nanoparticles for controlled delivery of paclitaxel. J Control Release 125: 96–106.
- Kumari A, Yadav SK, Pakade YB, Singh Bi, Yadav SC (2010) Development of biodegradable nanoparticles for delivery of quercetin. Colloids Surf B Biointerfaces 80: 184–92.
- JB Naik, Mokale VJ, Shevalkar GB, Patil KV, JS Patil, et al. (2013) Formulation and evaluation of poly (L-lactide-co--caprolactone) loadedgliclazide biodegradable nanoparticles as a control release carrier. Int J Drug Deliv 5: 300-8.
- Yadava SK, Patil JS, Mokale VJ, Naik JB (2014) Sodium alginate/HPMC/liquid paraffin emulsified (o/w) gel beads, by factorial design approach; and in vitro analysis. J Sol-Gel Sci Technol 71: 60-8.
- Khairnar G, Mokale V, Naik J (2014) Formulation and development of nateglinide loaded sustained release ethyl cellulose microspheres by O/W solvent emulsification technique. J Pharma Investig doi: 10.1007/s40005-014-0118-3.
- Mokale V, Naik J, Sutar Y, Khairnar G (2014) Chitosan reinforced alginate controlled release beads of losartan potassium: design, formulation and in vitro evaluation. J Pharma Investig doi: 10.1007/s40005-014-0122-7.
- Grant N, Zhang H (2011) Poorly water-soluble drug nanoparticles via an emulsion-freeze-drying approach. J Colloid Int Sci 356: 573–8.
- Shekunov BY, Chattopadhyay P, Seitzinger J, Huff R (2006) Nanoparticles of Poorly Water-Soluble Drugs Prepared by Supercritical Fluid Extraction of Emulsions. Pharm Research 23: 196-204.
- Kipp JE (2004) The role of solid nanoparticle technology in the parenteral delivery of poorly water-soluble drugs. Int J Pharm 284: 109–122.
- Deshmukh RK, Naik JB (2013) Diclofenac Sodium-Loaded Eudragit® MicrospheresOptimizationUsing Statistical Experimental Design. J Pharm Innov 8: 276–87.
- Deshmukh RK, Naik JB (2014) Aceclofenac microspheres: Quality by design approach. Mater Sci Eng C Mater Biol Appl 36: 320–8.
- Naik JB, Lokhande AB, Mishra S, Kulkarni RD (2012) Development of sustained release micro/nanoparticles using different solvent emulsification technique: A review. Int J Pharm Bio Sci 3: 573-90.
- Avachat AM, GujarKN, Wagh KV (2013) Carbohydrate Polymers 91: 537–542.
- Costa P, Lobo JMS (2001) Modelling and comparison of dissolution profiles. Euro J Pharm Sci 13: 123–33.
- Sharma R, Walker RB, Pathak K (2011) Evaluation of the Kinetics and Mechanism of Drug Release from Econazole nitrate Nanosponge Loaded Carbapol Hydrogel. Ind J Pharm Edu Res 45.
- Patil JS, Yadav SK, Mokale VJ, Naik JB (2013) Development of surfactant free nanoparticles by a single Emulsion high pressure homogenization technique and effect of Formulation parameters on the drug entrapment and release. Int J Pharm 3: 843-52.
- Maulvi FA, Dalwadi SJ, Thakkar VT, Sonic GT, Goheld MC, et al. (2011) Improvement of dissolution rate of aceclofenac by solid dispersion technique. Powder Tech 207: 47–54.
- Newman A, Knipp G, Zogra G (2012) Assessing the performance of amorphous solid dispersions. J Pharm Sci 101: 1355–77.
- Tadros MI (2010) Controlled-release effervescent floating matrix tablets of ciprofloxacin hydrochloride: Development, optimization and in vitro-in vivo evaluation in healthy human volunteers. Euro J Pharm Biopharm 74: 332-9.
- Verma U, Naik JB, Mokale VJ (2014) Preparation of Freeze-dried Solid Dispersion Powder using Mannitol to Enhance Solubility of Lovastatin and Development of Sustained Release Tablet Dosage Form. American J Pharm Sci Nanotech 1: 11-26.
- Shoaib MH, Tazeen J, Merchant HA, Yousuf RI (2006) Evaluation of drug release kinetics from ibuprofen matrix tablets using HPMC. Pak J Pharm Sci 19: 119-124.
- Author Information
-
Mokale VJ, Naik JB*, Verma U, Patil JS and Yadava SK Department of Pharmaceutical Technology, University Institute of Chemical Technology, North Maharashtra University, Maharashtra, India - Article history
-
Received Accepted Published 16 June 2014 24 June 2014 18 August 2014 - View Pdf
-

- Tables and Figures
-
IMAGE 1 IMAGE 2 IMAGE 3 IMAGE 4 IMAGE 5 IMAGE 6 IMAGE 7 TABLE 1 TABLE 2 TABLE 3 TABLE 4 - Citation
-
Mokale VJ, Naik JB, Verma U, Patil JS, Yadava SK (2014) Preparation and characterization of Biodegradable Glimepiride Loaded PLA Nanoparticles by o/w Solvent Evaporation method using high pressure homogenizer: A factorial design approach. SAJ Pharma Pharmacol 1: 104. doi: 10.18875/2375-2262.1.104