Study on Rice Breeding Resistant to Brown Planthoper (Nilaparvata Lugens Stål) in Mekong Delta, Vietnam
The brown planthopper (BPH) is the most serious pest threat to rice production throughout Asia. The materials including 23 varieties and 13 of BC1F1 populations were evaluated for the screening of BPH. The germplasm was screened for BPH following standard of IRRI in the greenhouse. Twenty-three (23) varieties also were used to evaluate the cluster diagram based on by phenotype. The phenotypic evaluation showed a clear distinction between resistant and susceptible type by clearly revealing moderately resistant types as well. From the total of 23 varieties screened, the results showed varieties with good resistance (OM6619, OM6062, OM5239, AS996, OM4498, OM4495). The marker VL3,4 linked to BPH resistance from the nine of BC1F1 combinations. The closely linked marker identified in this study should be useful in marker-assisted breeding to produce BPH resistant cultivars. The combined use of phenotypic and genotypic evaluation methods can improve the productivity of marker-assisted selection and application for resistant genotypes for crop development by rice breeders.
Keywords: Brown Planthopper; Rice; Screening; Marker; Resistance
Rice (Oryza sativa L.) is one of the important food crops, which feed more than one-third of the world’s population [1,2]. In Vietnam, rice is not only a food crop but also a major contributor to the agricultural sector. During the growth process, the rice is subjected to many diseases and insect outbreaks that reduce the yield and quality of rice. Brown planthopper (BPH) is one of the causing general losses in rice production in the tropical, subtropical and temperate areas in Asia and the main reason of rice production lost is an outbreak of brown planthopper in Mekong Delta, Southern Vietnam from 2005 to 2006 [3-6]. Four biotypes BPH on rice have been known [7]. Biotype 1 was distributed in South-east Asia, while biotype 2 was originated in the Indonesia and Vietnam [8]. Biotype 3 occurs at IRRI and Japan whereas biotype 4 is found only in South Asia [5]. In addition to directly causing “hopper burn”, BPH is also known to transmit different serious viral diseases as the ragged stunt and grassy stunt virus [9,10]. Although the timely application of pesticides provides effective control, large-scale chemical control is difficult and expensive. The use of genetic resistance rice varieties is the most economical and efficient and environmentally sound strategy for controlling the BPH [11,12]. Generally, the markers are validated in fine mapping studies. As said by the thought of DNA marker-assisted selection, the target genes can be identified in a segregating the population at any plant growth stage based on linked molecular marker. Therefore, it is positive to identify BPH resistance genes from varied sources and combine them into rice cultivars. Identification of new donors is a continuous process to breed new BPH resistant varieties by the use of modern molecular tools. The objective of the present study was to evaluate BPH resistance of breeding materials and selection genetic variability among different populations based on molecular approach method.
A total of 23 varieties and 13 of BC1F1 populations were screened for resistance to BPH. Ptb33 and TN1 were chosen as resistant and susceptible control varieties, respectively.
One BPH population, which was collected from the rice field in Can tho city, was used to evaluate the BPH resistance. The insects were reared on rice cultivar TN1. The adults were confined on 35day-old potted plants of TN1 placed in oviposition cages (50 x 50 x 60 cm) having aluminum frames, glass top, door and wire mesh sidewalls. The oviposition insects were removed three days later and plants with eggs were taken out of cages, placed in separate cages for the nymphs to emerge. The emerged nymphs were then transferred to the 15- day – old TN 1 seedlings raised in the germination trays, which in turn were placed in galvanized iron trays containing a 5cm depth of water to increase humidity and to avoid watering daily. The raised seedlings were changed as and when necessary. Using this technique, a continuous culture of the BPH was maintained during the period of study.
Phenotypic screening for BPH resistance: Screening of parents and segregating progenies for resistance to BPH were done using seed box screening test (SSST) in greenhouse conditions. The experiment was conducted at a temperature of 28 to 30 °C and relative humidity of 70 to 80 percent in the controlled greenhouse. A brief description of the procedure is provided below. The seeds were pre-soaked and sown in rows (2 cm interval) in 40 x 60 x 10 cm seed boxes along with checks, PTB33 (resistant) and TN1 (susceptible). A row of 30 seedlings was maintained per variety rice. Seven – ten-day-old seedlings were infested with first instar nymphs at the rate of 6-8 per seedling. Approximately one week after infestation ‘hopperburn’ symptom was observed.
When all TN1 plants dead, the damage rating based on scoring system proposed by International Rice Research Institute was used to rate each seedling, 0 = no injury; 1 = very slight injury; 3 = first and 2nd leaves of most plant partially yellowing; 5 = pronounced yellowing and stunting or about 10-25% of the plants wilting or dead and remaining plants severely stunted or dying; 7 = more than half of the plants died, the plant was still alive; 9 = all plants dead [13].
DNAs for PCR analysis were prepared using a simplified mini scale procedure [14]. A piece of young rice leaf (2 cm) was collected and placed in a labeled 1.5 ml centrifuge tube in ice. The leaf was ground using a polished glass rod in a well of a Spot Test Plate (Thomas Scientific) after adding 400 μl of extraction buffer (50 mM Tris-HCl pH 8.0, 25mM EDTA, 300mM NaCl and 1% SDS). Grinding was done until the buffer turned green which is an indication of cell breakage and release of chloroplasts and cell contents. Another 400 μl of the extraction buffer was added and mixed into the well by pipetting. Around 400 μl of the lysate was transferred to the original tube of the leaf sample. The lysate was deproteinized using 400 μl of chloroform. The aqueous supernatant was transferred to a new 1.5 ml tube and DNA precipitated using absolute ethanol. DNA was air-dried and resuspended in 50 μl of TE buffer (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0). An aliquot of 1μl is sufficient for PCR analysis. The remaining DNA was stored in -20 °C for any later use.
The PCR reaction mixture contained 20-50 nanogram (ng) template DNA, 50ng of each primer, 0.05 mM dNTP’s, 1X PCR buffer (10mM Tris pH 8.4, 50mM KCl, 1.8mM MgCl2 and 0.01 mg/ml gelatin) and 1 unit of Taq DNA polymerase in a total volume of 20 μl. Template DNA was initially denatured at 94 °C for 5 minutes followed by 35 cycles of PCR amplification using the following parameters: 30 sec denaturation of 94 °C, 30 sec primer annealing at 55 °C or 60 °C and 1 min primer extension at 72 °C. Completion of primer extension was allowed by final 5 min incubation at 72 °C.
Agarose gel electrophoresis: The PCR products were run on 3 percent gels to verify amplification. The gel was electrophoresed in 1X TAE at 100V for 50min. The gel was then stained with ethidium bromide solution (1μg /ml) for 15 min. and de-stained with water and observed on a UV transilluminator. This exercise was made just to confirm whether the amplification is satisfactory or not before the PCR products are separated on 5% polyacrylamide gel for better resolution.
PCR products were separated on 6.0% polyacrylamide denaturing gels and isualized by means of silver staining (or 3% agarose denaturing gels and isualized by means of ethidium bromide staining).
Scoring of the gel: The amplified fragments were scored as ‘1’ for the presence, ‘0’ for the absence of the alleles from higher to lower molecular weight products.
Primers were designed based on sequences obtained from the automated sequencing experiments. The available PCR marker for BPH is based on the linked SSR marker VL3,4. The primer was designed based on DNA sequences derived by manual sequencing from both ends of SSR clone. This primer PCR product needed to be digested with HinfI. When monomorphic bands were obtained, several restriction enzymes were tried on the PCR products to generate a PCR based. In this study, SSR marker VL3,4 linked to BPH resistance was used for analysis in the BC1F1 generation by the enzyme Hinf1 showed marked polymorphism with molecular size is 800bp - 1000bp (infection) and 1000bp-1100bp (resistance).
Forward primer: 5’CCGAGACAAGCTGTGACGCT3’
Reverse primer: 5’ACATTAGGGCCATGTTCCTCG3’
The experimental design used was completely randomized design (CRD) with three replications. The scores obtained in all varieties in the damaged rating analysis. The raw values of 23 varieties were subjected to cluster using the computer software NTSYS pcv2.02i [15].
The BPH has expected great importance in rice and often occurs as the most damaging pest of rice in many countries. The first directly evaluate host resistance by measuring the degree of damage following BPH infestation. The results of the phenotypic response of varieties to brown planthopper screening at seedling stage indicated varied phenotypic responses (Table 1). PTB33 widely used as the resistant check for BPH by rice breeders consisting of Bph2 and Bph3 genes scored 3, and TN1 showed a score of 9 as a susceptible check [8]. The seedling resistance of 23 varieties on BPH was examined in this study. High resistance (Score 1) was shown by OM6619, followed by four varieties (OM6620, OM6062, OM5239, and Ptb33 exhibited moderate degrees of resistance (scoring 3) TN1 was found to be highly susceptible (score 9) and 17 varieties susceptible (scoring 5). Conclusively, evaluation of germplasm for resistance genes plays a major role in the selection of parental lines and development of new breeding material. The resistant varieties found could provide a good basis for breeding new resistant varieties in future breeding programs in Vietnam.
Identification of donors for resistance and efficient screening techniques for evaluating breeding lines plays a crucial role to transfer BPH resistance genes into high yielding popular varieties and a high level of genetic diversity reduces the risk of widespread epidemics of pests and diseases [16,17]. This strategy is very similar to what has been performed for analysis of varieties using gene evaluation.
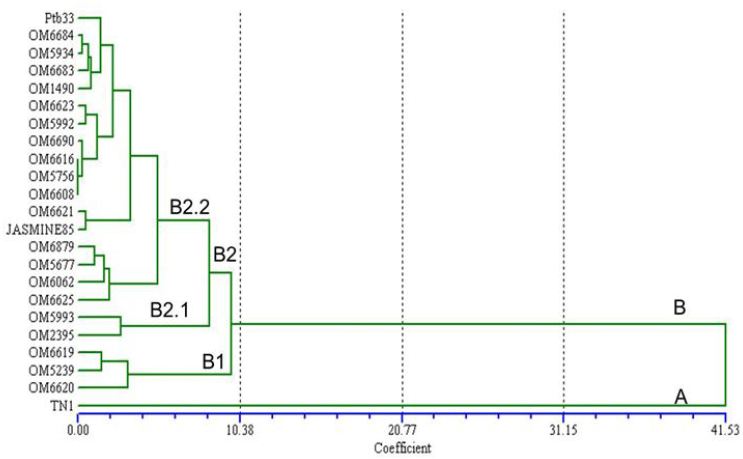
The same association relatedness is depicted by the neighbor-joining tree (Figure 1). The 23 varieties were categorized into two clusters A and B. Cluster A included TN1 showing susceptible, cluster B is comprised of two sub-clusters B1 and B2. Sub-cluster B1 included all moderately resistant varieties and sub-cluster B2 comprised of two sub-groups B2.1 and B2.2. All the susceptible varieties were grouped into cluster B2. The tree showed a close relationship between resistance and moderately resistant genotypes based on BPH resistance phenotype analysis. However, phenotyping method could clearly differentiate rice genotypes categorized as resistant and moderately resistant genotypes. The seedling resistance of 23 varieties on BPH was examined in this study
Use of marker and phenotype information together offers an effective tool to the breeders in selecting parents for various breeding programs. In total, 13 of BC1F1 populations and the 9 parents were screened for BPH resistance based on the standard evaluation system of BPH damage to rice (Table 2).
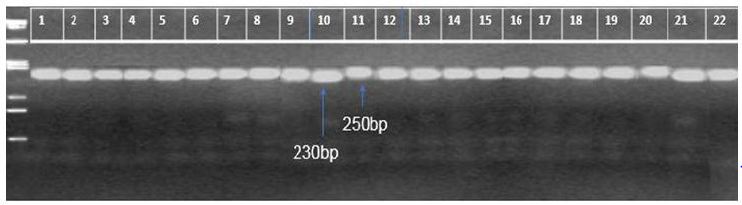
The donor parents AS996, OM4498, ST3, OM5930, and OM4495 expressed moderately resistance to the BPH population used in this study, while TN1 and IR64 and OM4900 were completely susceptible to the BPH. The BC1 of 10 populations were resistant, whereas the three combinations were moderately resistant to the BPH (Table 2). In Figure 2, the progenies having a resistant allele (250 bp) of marker VL3,4 represent BPH resistance. It is noted that thirteen combinations (1, 2, 3, 4, 5, 6, 7, 8, 11, 12, 13, 14, and 15) were resistance with BPH. Marker VL3,4 linked resistance gene Bph-4 of Babawee (biotype 4) in TN1/ Babawee population (8). So, this marker linked to resistance gene in this study would also be useful for breeding programs. The Bph-4 gene was first identified in rice cultivar Babawee [18]. The VL3,4 marker linked to the Bph-4 locus in this study would also be useful for MAS in BPH-resistant breeding programs.
Another important aspect relates to using phenotype information together offers an effective tool to the breeders in selecting parents for various breeding programs. Among all the thirteen combinations and their parent's verities (Table 2). TN1 has shown highly susceptible (scoring 9) to BPH whereas, insects on Ptb33 showed resistance (scoring 3). Nine combinations showed high resistance (scoring 1) and three combinations showed high to moderate resistance.
The results in Table 3 showed that the resistant and susceptible of BC1F1 combinations showed a 15: 1 segregation ratio belong to 6 populations consist of 2, 3,4, 6, 8 and 11 (χ2 =0.44, χ2 = 1.29, χ2 =0.18, χ2 = 0.44, χ2 = 0.91, and χ2 = 2.21, respectively). The segregation of phenotypes of combinations did not follow Mendel's theory due to the impact of environmental conditions combined with the pressure of different aphids. The data provided very useful information for formulating strategies for improving BPH resistance in rice breeding programs, which have now been implemented.
Based on the results of genotyping and phenotypic analysis, the majority of varieties or combinations exhibited resistance to brown backed rice planthopper, both of which showed that the combinations and cultivars had a good resistance to brown backed rice planthopper, it is necessary to further select and develop these varieties and combinations. However, there are still some breeds showing infection, so they need to be repaired.
The results showed that OM6619, OM6062, OM5239, AS996, OM4498, OM4495 were able to produce good resistance to BPH backed rice nests. Filtered 9 resistance combinations (1OM6602 / OM4900 // OM6602, OM5239 / OM4900 // OM5239, OM6620 / OM4900 // OM6602, OM6619 / OM4495 // OM6619, OM6602 / OM4498 // OM6602, DBTD / ST3 // DBTDT, DBTDT / ST3 // DBTDT, AS996 / Basmati370 // AS996, OM4498 / OM4495 // OM4498) and expression resistance when genotyping demonstrates that these combinations carry valuable gene sources and need further investigation to reveal this gene source is used for breeding.
In addition, TN1 is a crossbreed for Babawee hybridization with Bph4 biotype 4 which received the resistance gene from the Babawee. SSR analysis of marker VL3,4 results in the detection of resistance and infection genes that result in the majority of combinations with resistant genes. Use of marker and phenotype information together offers an effective tool to the breeders in selecting parents for various breeding programs. Due to the limited use of primers, it is not possible to determine the exact resistance gene. The genetic information generated in this research will be useful in rice breeding programmes to provide varieties with robust resistance to BPH.